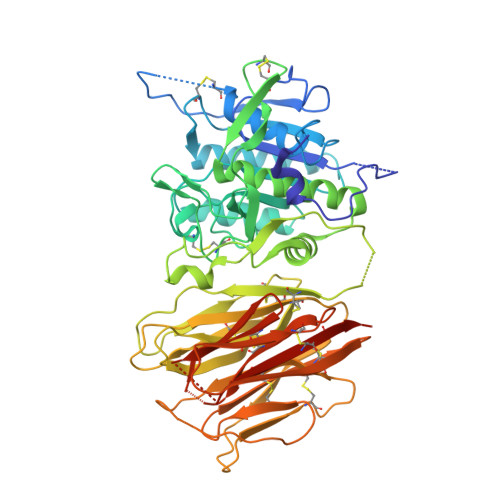
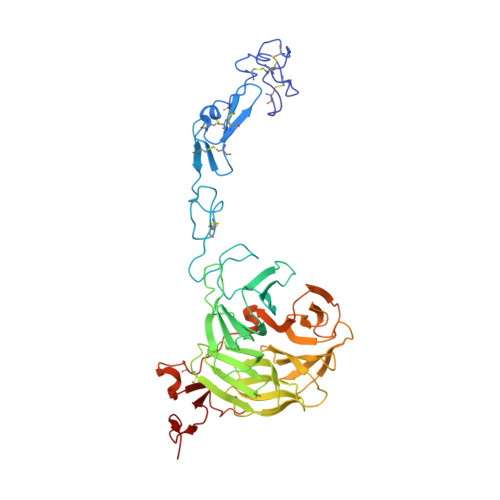
Mechanistic implications for LDL receptor degradation from the PCSK9/LDLR structure at neutral pH.
Lo Surdo, P., Bottomley, M.J., Calzetta, A., Settembre, E.C., Cirillo, A., Pandit, S., Ni, Y.G., Hubbard, B., Sitlani, A., Carfi, A.(2011) EMBO Rep 12: 1300-1305
- PubMed: 22081141
- DOI: https://doi.org/10.1038/embor.2011.205
- Primary Citation of Related Structures:
3P5B, 3P5C - PubMed Abstract:
The protein PCSK9 (proprotein convertase subtilisin/kexin type 9) is a key regulator of low-density lipoprotein receptor (LDLR) levels and cardiovascular health. We have determined the crystal structure of LDLR bound to PCSK9 at neutral pH. The structure shows LDLR in a new extended conformation. The PCSK9 C-terminal domain is solvent exposed, enabling cofactor binding, whereas the catalytic domain and prodomain interact with LDLR epidermal growth factor(A) and β-propeller domains, respectively. Thus, PCSK9 seems to hold LDLR in an extended conformation and to interfere with conformational rearrangements required for LDLR recycling.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, IRBM P. Angeletti, Via Pontina Km 30.600, Pomezia, Rome I-00040, Italy.