Crystal Structure of human 5'-deoxy-5'-methyladenosine phosphorylase
Ho, M., Guan, R., Almo, S.C., Schramm, V.L.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| S-methyl-5'-thioadenosine phosphorylase | 283 | Homo sapiens | Mutation(s): 0 Gene Names: MTAP, MSAP EC: 2.4.2.28 | 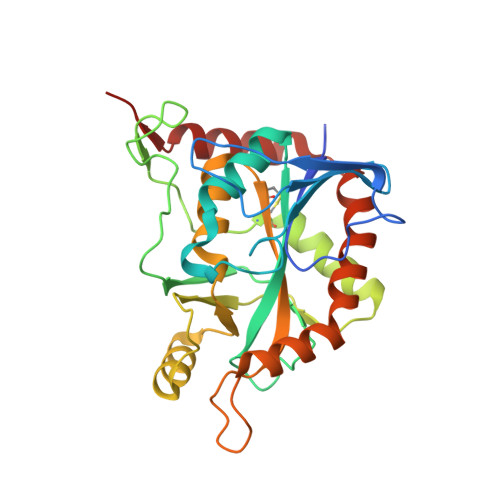 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q13126 (Homo sapiens) Explore Q13126 Go to UniProtKB: Q13126 | |||||
PHAROS: Q13126 GTEx: ENSG00000099810 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q13126 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 4CT Query on 4CT | C [auth A], D [auth B] | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-{[(4-chlorophenyl)sulfanyl]methyl}pyrrolidin-3-ol C18 H20 Cl N5 O S MZZBHZOHYGEGEE-DOMZBBRYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 121.539 | α = 90 |
| b = 121.539 | β = 90 |
| c = 87.661 | γ = 120 |
| Software Name | Purpose |
|---|---|
| DENZO | data reduction |
| SCALEPACK | data scaling |
| MOLREP | phasing |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |