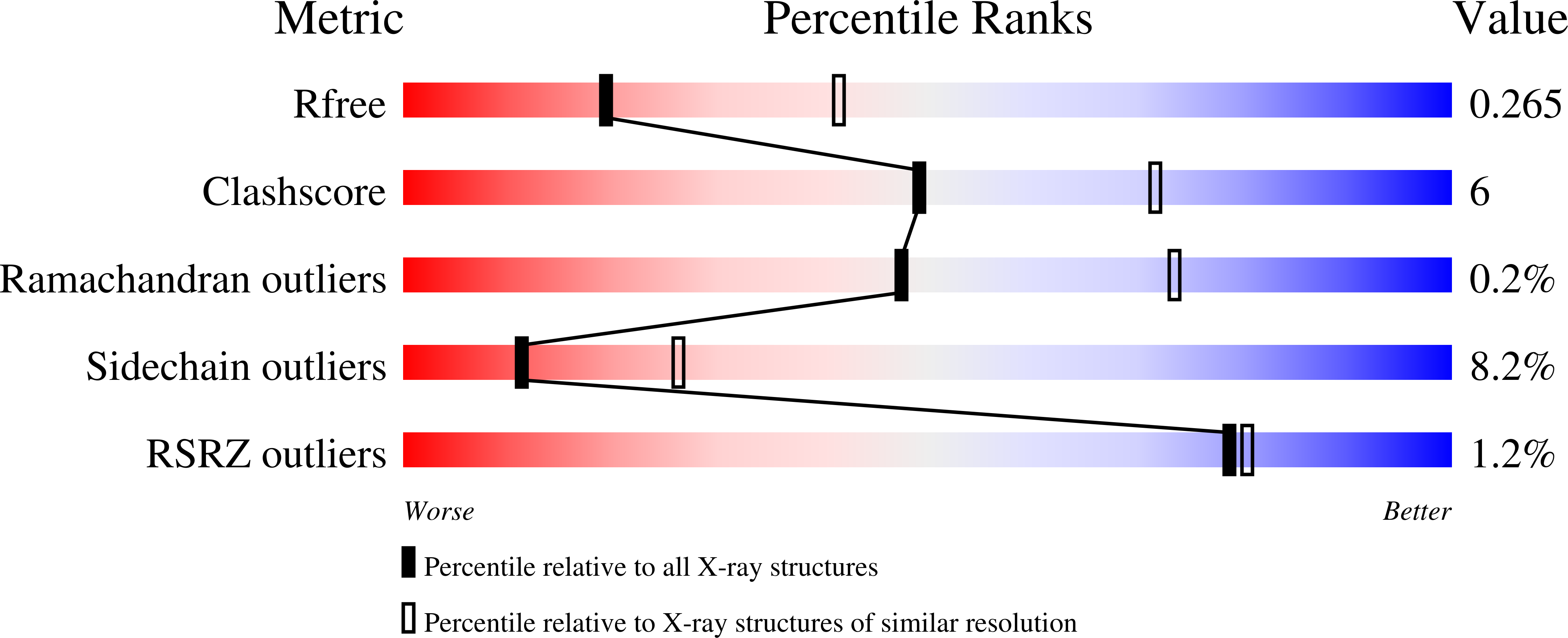
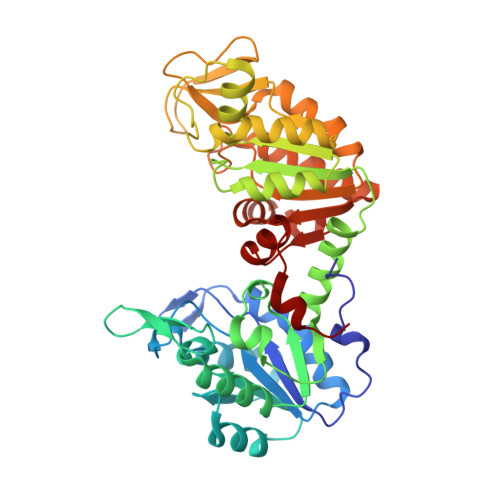
Crystal structure of Plasmodium falciparum phosphoglycerate kinase: Evidence for anion binding in the basic patch.
Smith, C.D., Chattopadhyay, D., Pal, B.(2011) Biochem Biophys Res Commun 412: 203-206
- PubMed: 21798238
- DOI: https://doi.org/10.1016/j.bbrc.2011.07.045
- Primary Citation of Related Structures:
3OZ7, 3OZA - PubMed Abstract:
3-Phosphoglycerate kinase (EC 2.7.2.3) is a key enzyme in the glycolytic pathway and catalyzes an important phosphorylation step leading to the production of ATP. The crystal structure of Plasmodium falciparum phosphoglycerate kinase (PfPGK) in the open conformation is presented in two different groups, namely I222 and P6(1)22. The structure in I222 space group is solved using MAD and refined at 3Å whereas that in P6(1)22A is solved using MR and refined at 2.7Å. I222 form has three monomers in asymmetric unit whereas P6(1)22 form has two monomers in the asymmetric unit. In both crystal forms a sulphate ion is located at the active site where ATP binds, but no Mg(2+) ion is observed. For the first time another sulphate ion is found at the basic patch where the 3-phosphate of 1,3-biphosphoglycerate normally binds. This was found in both chains of P6(1)22 form but only in chain A of I222 form.
Organizational Affiliation:
University of Alabama at Birmingham, Birmingham, AL 35294, USA.