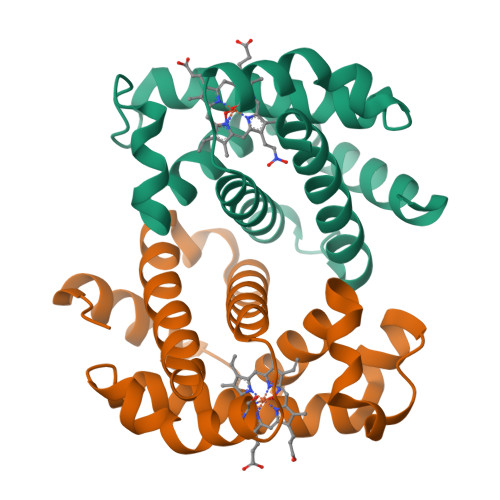
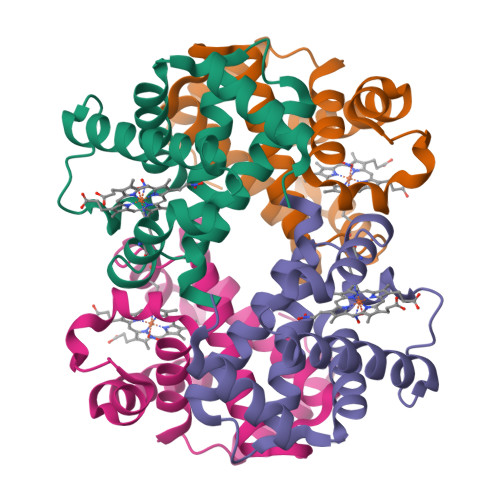
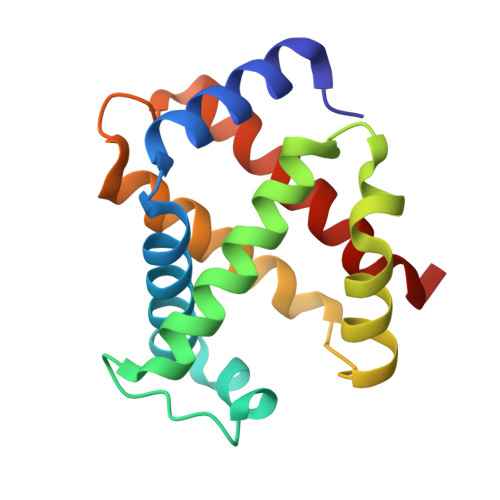
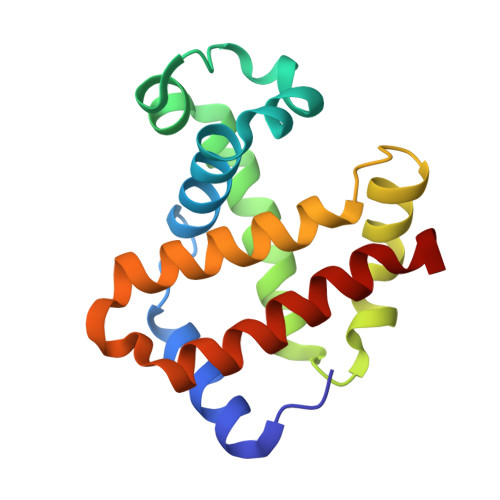
Crystallographic Trapping of Heme Loss Intermediates during the Nitrite-Induced Degradation of Human Hemoglobin.
Yi, J., Thomas, L.M., Musayev, F.N., Safo, M.K., Richter-Addo, G.B.(2011) Biochemistry 50: 8323-8332
- PubMed: 21863786
- DOI: https://doi.org/10.1021/bi2009322
- Primary Citation of Related Structures:
3ONZ, 3OO4, 3OO5 - PubMed Abstract:
Heme is an important cofactor in a large number of essential proteins and is often involved in small molecule binding and activation. Loss of heme from proteins thus negatively affects the function of these proteins but is also an important component of iron recycling. The characterization of intermediates that form during the loss of heme from proteins has been problematic, in a large part, because of the instability of such intermediates. We have characterized, by X-ray crystallography, three compounds that form during the nitrite-induced degradation of human α(2)β(2) hemoglobin (Hb). The first is an unprecedented complex that exhibits a large β heme displacement of 4.8 Å toward the protein exterior; the heme displacement is stabilized by the binding of the distal His residue to the heme Fe, which in turn allows for the unusual binding of an exogenous ligand on the proximal face of the heme. We have also structurally characterized complexes that display regiospecific nitration of the heme at the 2-vinyl position; we show that heme nitration is not a prerequisite for heme loss. Our results provide structural insight into a possible pathway for nitrite-induced loss of heme from human Hb.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of Oklahoma, Norman, Oklahoma 73019, United States. yijun@ou.edu