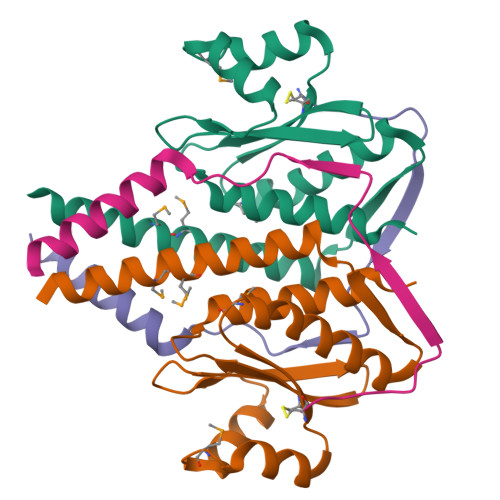
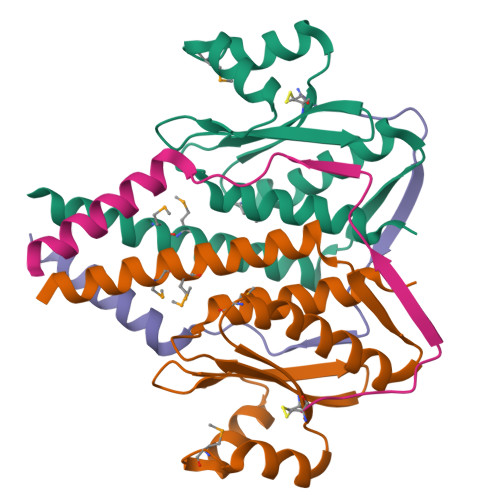
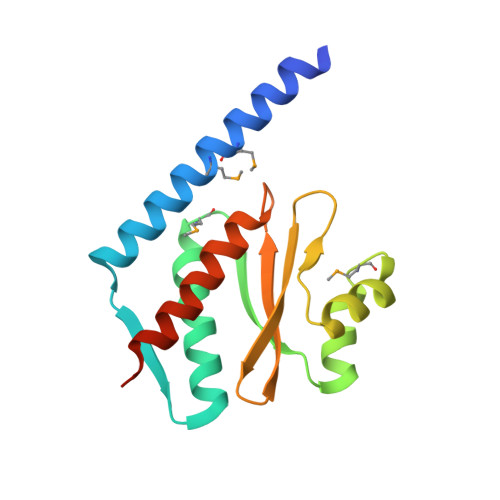
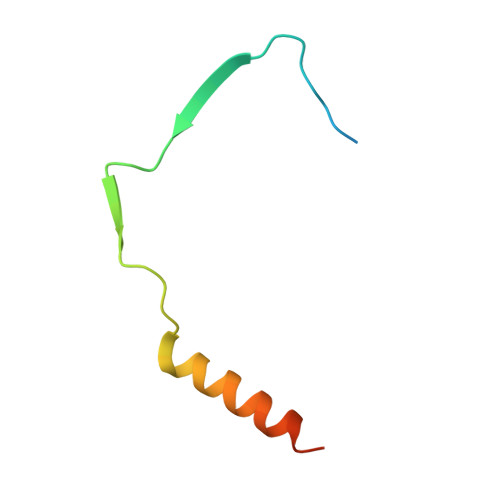
The structure and interactions of SpoIISA and SpoIISB, a toxin-antitoxin system in Bacillus subtilis.
Florek, P., Levdikov, V.M., Blagova, E., Lebedev, A.A., Skrabana, R., Resetarova, S., Pavelcikova, P., Barak, I., Wilkinson, A.J.(2010) J Biological Chem 161: 750-756
- PubMed: 21147767
- DOI: https://doi.org/10.1074/jbc.M110.172429
- Primary Citation of Related Structures:
3O6Q - PubMed Abstract:
Spore formation in Bacillus subtilis begins with an asymmetric cell division, following which differential gene expression is established by alternative compartment-specific RNA polymerase σ factors. The spoIISAB operon of B. subtilis was identified as a locus whose mutation leads to increased activity of the first sporulation-specific sigma factor, σ(F). Inappropriate spoIISA expression causes lysis of vegetatively growing B. subtilis cells and Escherichia coli cells when expressed heterologously, effects that are countered by co-expression of spoIISB, identifying SpoIISA-SpoIISB as a toxin-antitoxin system. SpoIISA has three putative membrane-spanning segments and a cytoplasmic domain. Here, the crystal structure of a cytoplasmic fragment of SpoIISA (CSpoIISA) in complex with SpoIISB has been determined by selenomethionine-multiwavelength anomalous dispersion phasing to 2.5 Å spacing, revealing a CSpoIISA(2)·SpoIISB(2) heterotetramer. CSpoIISA has a single domain α/β structure resembling a GAF domain with an extended α-helix at its N terminus. The two CSpoIISA protomers form extensive interactions through an intermolecular four-helix bundle. Each SpoIISB chain is highly extended and lacking tertiary structure. The SpoIISB chains wrap around the CSpoIISA dimer, forming extensive interactions with both CSpoIISA protomers. CD spectroscopy experiments indicate that SpoIISB is a natively disordered protein that adopts structure only in the presence of CSpoIISA, whereas surface plasmon resonance experiments revealed that the CSpoIISA·SpoIISB complex is stable with a dissociation constant in the nanomolar range. The results are interpreted in relation to sequence conservation and mutational data, and possible mechanisms of cell killing by SpoIISA are discussed.
Organizational Affiliation:
Institute of Molecular Biology, Slovak Academy of Sciences, 845 51 Bratislava 45, Slovakia.