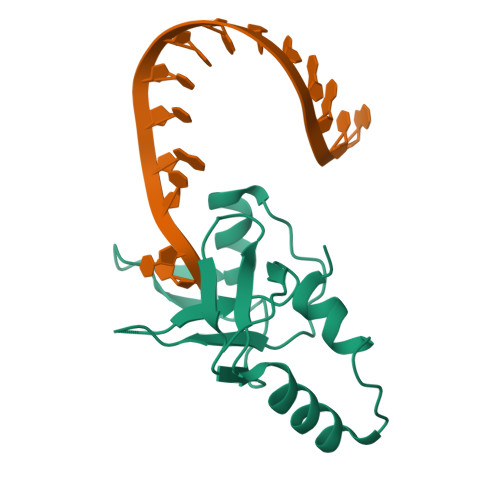
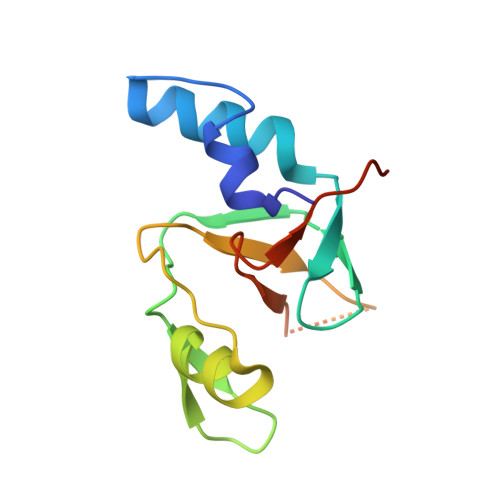
Inaugural Article: Structural basis for piRNA 2'-O-methylated 3'-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains.
Tian, Y., Simanshu, D.K., Ma, J.B., Patel, D.J.(2011) Proc Natl Acad Sci U S A 108: 903-910
- PubMed: 21193640
- DOI: https://doi.org/10.1073/pnas.1017762108
- Primary Citation of Related Structures:
3O3I, 3O6E, 3O7V, 3O7X - PubMed Abstract:
Argonaute and Piwi proteins are key players in the RNA silencing pathway, with the former interacting with micro-RNAs (miRNAs) and siRNAs, whereas the latter targets piwi-interacting RNAs (piRNAs) that are 2'-O-methylated (2(')-OCH(3)) at their 3' ends. Germline-specific piRNAs and Piwi proteins play a critical role in genome defense against transposable elements, thereby protecting the genome against transposon-induced defects in gametogenesis and fertility. Humans contain four Piwi family proteins designated Hiwi1, Hiwi2, Hiwi3, and Hili. We report on the structures of Hili-PAZ (Piwi/Argonaute/Zwille) domain in the free state and Hiwi1 PAZ domain bound to self-complementary 14-mer RNAs (12-bp + 2-nt overhang) containing 2(')-OCH(3) and 2'-OH at their 3' ends. These structures explain the molecular basis underlying accommodation of the 2(')-OCH(3) group within a preformed Hiwi1 PAZ domain binding pocket, whose hydrophobic characteristics account for the preferential binding of 2(')-OCH(3) over 2'-OH 3' ends. These results contrast with the more restricted binding pocket for the human Ago1 PAZ domain, which exhibits a reverse order, with preferential binding of 2'-OH over 2(')-OCH(3) 3' ends.
Organizational Affiliation:
Structural Biology Program, Memorial Sloan-Kettering Cancer Center, New York, NY 10065, USA.