Mechanistic, mutational, and structural evaluation of a taxus phenylalanine aminomutase.
Feng, L., Wanninayake, U., Strom, S., Geiger, J., Walker, K.D.(2011) Biochemistry 50: 2919-2930
- PubMed: 21361343
- DOI: https://doi.org/10.1021/bi102067r
- Primary Citation of Related Structures:
3NZ4 - PubMed Abstract:
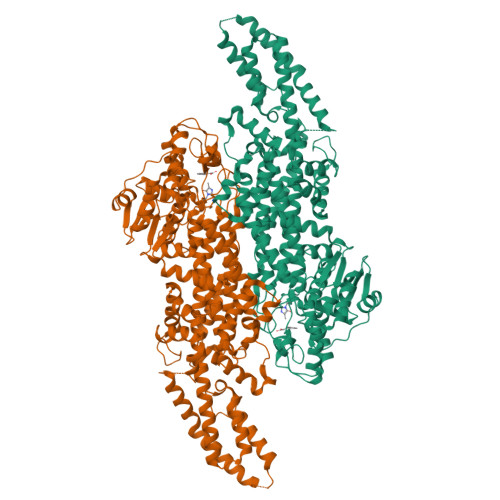
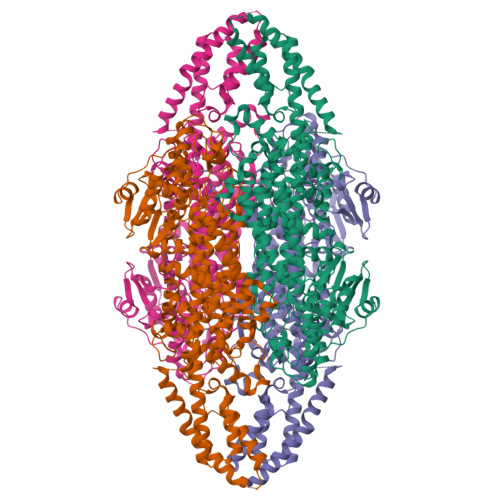
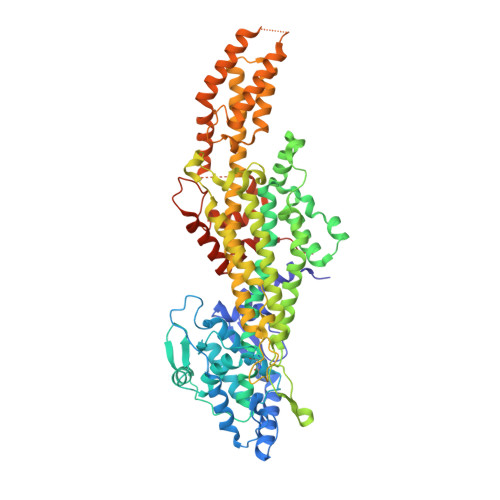
The structure of a phenylalanine aminomutase (TcPAM) from Taxus canadensis has been determined at 2.4 Å resolution. The active site of the TcPAM contains the signature 4-methylidene-1H-imidazol-5(4H)-one prosthesis, observed in all catalysts of the class I lyase-like family. This catalyst isomerizes (S)-α-phenylalanine to the (R)-β-isomer by exchange of the NH2/H pair. The stereochemistry of the TcPAM reaction product is opposite of the (S)-β-tyrosine made by the mechanistically related tyrosine aminomutase (SgTAM) from Streptomyces globisporus. Since TcPAM and SgTAM share similar tertiary- and quaternary-structures and have several highly conserved aliphatic residues positioned analogously in their active sites for substrate recognition, the divergent product stereochemistries of these catalysts likely cannot be explained by differences in active site architecture. The active site of the TcPAM structure also is in complex with (E)-cinnamate; the latter functions as both a substrate and an intermediate. To account for the distinct (3R)-β-amino acid stereochemistry catalyzed by TcPAM, the cinnamate skeleton must rotate the C1-Cα and Cipso-Cβ bonds 180° in the active site prior to exchange and rebinding of the NH2/H pair to the cinnamate, an event that is not required for the corresponding acrylate intermediate in the SgTAM reaction. Moreover, the aromatic ring of the intermediate makes only one direct hydrophobic interaction with Leu-104. A L104A mutant of TcPAM demonstrated an ∼1.5-fold increase in kcat and a decrease in KM values for sterically demanding 3'-methyl-α-phenylalanine and styryl-α-alanine substrates, compared to the kinetic parameters for TcPAM. These parameters did not change significantly for the mutant with 4'-methyl-α-phenylalanine compared to those for TcPAM.
Organizational Affiliation:
Department of Chemistry, Michigan State University, East Lansing, Michigan 48824, United States.