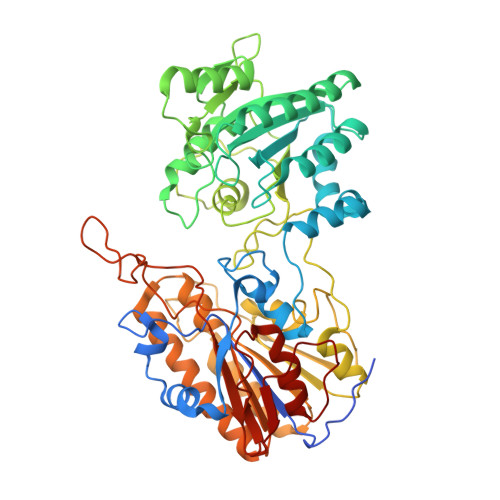
Crystal structure of phosphoglycerate mutase from Trypanosoma brucei.
Mercaldi, G.F., Pereira, H.M., Cordeiro, A.T., Michels, P.A., Thiemann, O.H.(2012) FEBS J 279: 2012-2021
- PubMed: 22458781
- DOI: https://doi.org/10.1111/j.1742-4658.2012.08586.x
- Primary Citation of Related Structures:
3NVL - PubMed Abstract:
Phosphoglycerate mutases (PGAMs) participate in both the glycolytic and the gluconeogenic pathways in reversible isomerization of 3-phosphoglycerate and 2-phosphoglycerate. PGAMs are members of two distinct protein families: enzymes that are dependent on or independent of the 2,3-bisphosphoglycerate cofactor. We determined the X-ray structure of the monomeric Trypanosoma brucei independent PGAM (TbiPGAM) in its apoenzyme form, and confirmed this observation by small angle X-ray scattering data. Comparing the TbiPGAM structure with the Leishmania mexicana independent PGAM structure, previously reported with a phosphoglycerate molecule bound to the active site, revealed the domain movement resulting from active site occupation. The structure reported here shows the interaction between Asp319 and the metal bound to the active site, and its contribution to the domain movement. Substitution of the metal-binding residue Asp319 by Ala resulted in complete loss of independent PGAM activity, and showed for the first time its involvement in the enzyme's function. As TbiPGAM is an attractive molecular target for drug development, the apoenzyme conformation described here provides opportunities for its use in structure-based drug design approaches. Database Structural data for the Trypanosoma brucei 2,3-bisphosphoglycerate-independent phosphoglycerate mutase (iPGAM) has been deposited with the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank under code 3NVL.
Organizational Affiliation:
Instituto de Física de São Carlos, Grupo de Cristalografia, Universidade de São Paulo, São Carlos, São Paulo, Brazil.