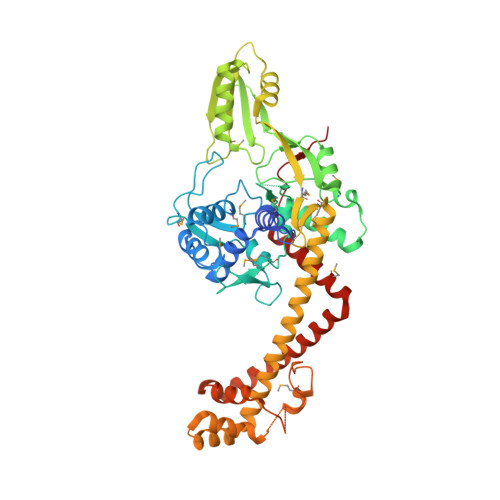
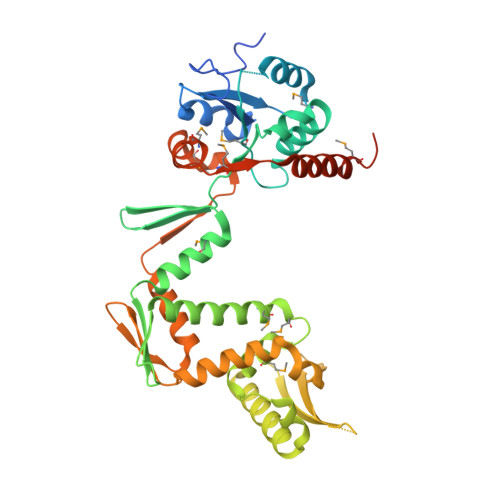
A domain insertion in Escherichia coli GyrB adopts a novel fold that plays a critical role in gyrase function.
Schoeffler, A.J., May, A.P., Berger, J.M.(2010) Nucleic Acids Res 38: 7830-7844
- PubMed: 20675723
- DOI: https://doi.org/10.1093/nar/gkq665
- Primary Citation of Related Structures:
3NUH - PubMed Abstract:
DNA topoisomerases manage chromosome supercoiling and organization in all forms of life. Gyrase, a prokaryotic heterotetrameric type IIA topo, introduces negative supercoils into DNA by an ATP-dependent strand passage mechanism. All gyrase orthologs rely on a homologous set of catalytic domains for function; however, these enzymes also can possess species-specific auxiliary regions. The gyrases of many gram-negative bacteria harbor a 170-amino acid insertion of unknown architecture and function in the metal- and DNA-binding TOPRIM domain of the GyrB subunit. We have determined the structure of the 212 kDa Escherichia coli gyrase DNA binding and cleavage core containing this insert to 3.1 Å resolution. We find that the insert adopts a novel, extended fold that braces the GyrB TOPRIM domain against the coiled-coil arms of its partner GyrA subunit. Structure-guided deletion of the insert greatly reduces the DNA binding, supercoiling and DNA-stimulated ATPase activities of gyrase. Mutation of a single amino acid at the contact point between the insert and GyrA more modestly impairs supercoiling and ATP turnover, and does not affect DNA binding. Our data indicate that the insert has two functions, acting as a steric buttress to pre-configure the primary DNA-binding site, and serving as a relay that may help coordinate communication between different functional domains.
Organizational Affiliation:
Department of Molecular and Cell Biology, California Institute for Quantitative Biosciences, University of California, Berkeley and Fluidigm Corporation, South San Francisco, CA 94080, USA. jmberger@berkeley.edu