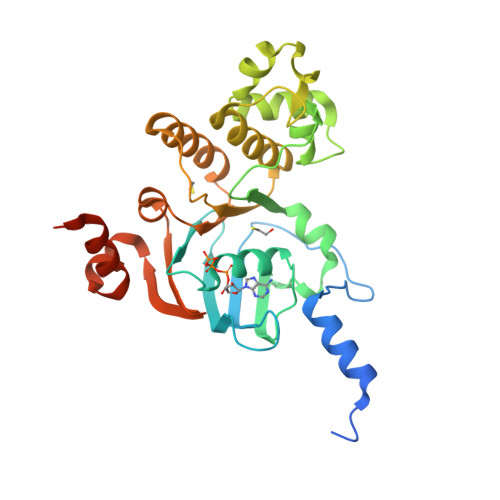
Structures of the nucleotide-binding domain of the human ABCB6 transporter and its complexes with nucleotides.
Haffke, M., Menzel, A., Carius, Y., Jahn, D., Heinz, D.W.(2010) Acta Crystallogr D Biol Crystallogr 66: 979-987
- PubMed: 20823549
- DOI: https://doi.org/10.1107/S0907444910028593
- Primary Citation of Related Structures:
3NH6, 3NH9, 3NHA, 3NHB - PubMed Abstract:
The human ATP-binding cassette (ABC) transporter ABCB6 is involved in haem-precursor transport across the mitochondrial membrane. The crystal structure of its nucleotide-binding domain (NBD) has been determined in the apo form and in complexes with ADP, with ADP and Mg(2+) and with ATP at high resolution. The overall structure is L-shaped and consists of two lobes, consistent with other reported NBD structures. Nucleotide binding is mediated by the highly conserved Tyr599 and the Walker A motif, and induces notable structural changes. Structural comparison with other structurally characterized NBDs and full-length ABC transporters gives the first insight into the possible catalytic mechanism of ABCB6 and the role of the N-terminal helix alpha(1) in full-length ABCB6.
Organizational Affiliation:
Helmholtz Zentrum für Infektionsforschung, Braunschweig, Germany.