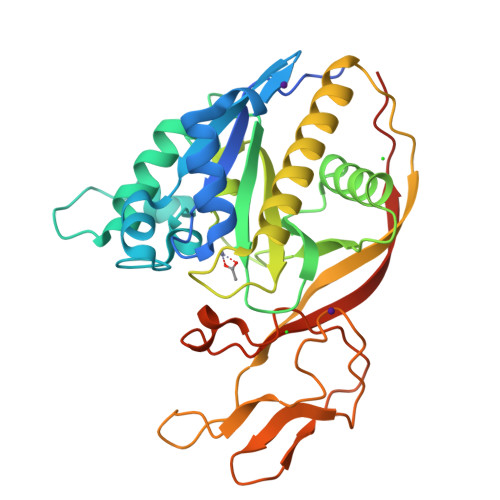
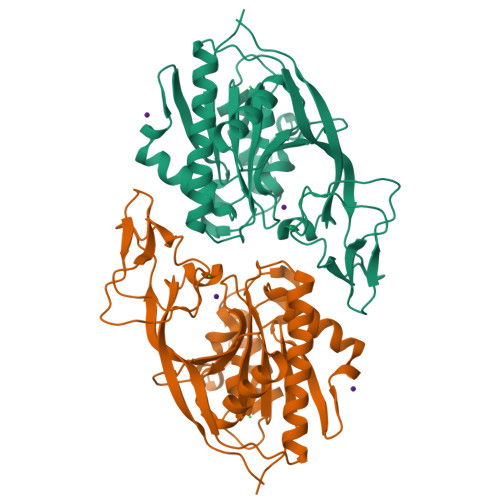
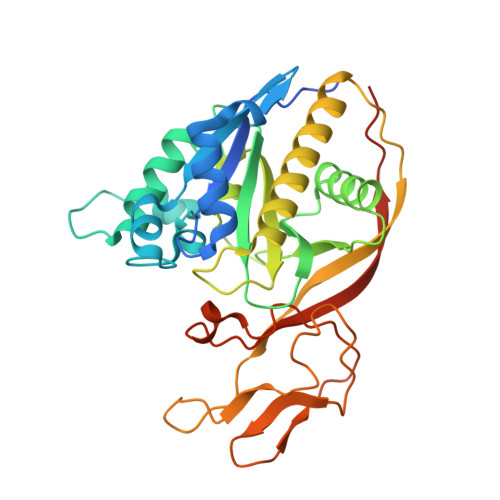
Structures of aminoacylase 3 in complex with acetylated substrates.
Hsieh, J.M., Tsirulnikov, K., Sawaya, M.R., Magilnick, N., Abuladze, N., Kurtz, I., Abramson, J., Pushkin, A.(2010) Proc Natl Acad Sci U S A 107: 17962-17967
- PubMed: 20921362
- DOI: https://doi.org/10.1073/pnas.1006687107
- Primary Citation of Related Structures:
3NFZ, 3NH4, 3NH5, 3NH8 - PubMed Abstract:
Trichloroethylene (TCE) is one of the most widespread environmental contaminants, which is metabolized to N-acetyl-S-1,2-dichlorovinyl-L-cysteine (NA-DCVC) before being excreted in the urine. Alternatively, NA-DCVC can be deacetylated by aminoacylase 3 (AA3), an enzyme that is highly expressed in the kidney, liver, and brain. NA-DCVC deacetylation initiates the transformation into toxic products that ultimately causes acute renal failure. AA3 inhibition is therefore a target of interest to prevent TCE induced nephrotoxicity. Here we report the crystal structure of recombinant mouse AA3 (mAA3) in the presence of its acetate byproduct and two substrates: N(α)-acetyl-L-tyrosine and NA-DCVC. These structures, in conjunction with biochemical data, indicated that AA3 mediates substrate specificity through van der Waals interactions providing a dynamic interaction interface, which facilitates a diverse range of substrates.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California, Los Angeles, CA 90095, USA.