Northeast Structural Genomics Consortium Target ClR27C
Kuzin, A., Abashidze, M., Seetharaman, J., Sahdev, S., Xiao, R., Ciccosanti, C., Wang, D., Everett, J.K., Nair, R., Acton, T.B., Rost, B., Montelione, G.T., Tong, L., Hunt, J.F.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Diguanylate cyclase/phosphodiesterase | 178 | Caldicellulosiruptor saccharolyticus DSM 8903 | Mutation(s): 0 Gene Names: Csac_0821 | 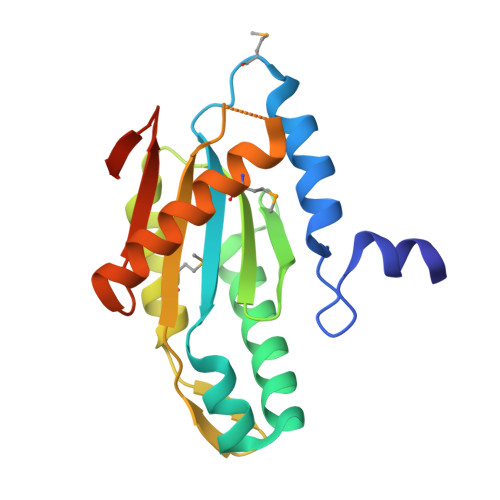 | |
UniProt | |||||
Find proteins for A4XHQ4 (Caldicellulosiruptor saccharolyticus (strain ATCC 43494 / DSM 8903 / Tp8T 6331)) Explore A4XHQ4 Go to UniProtKB: A4XHQ4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A4XHQ4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A, B | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 51.546 | α = 90 |
| b = 60.946 | β = 94.95 |
| c = 56.173 | γ = 90 |
| Software Name | Purpose |
|---|---|
| CNS | refinement |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| SnB | phasing |
| REFMAC | refinement |