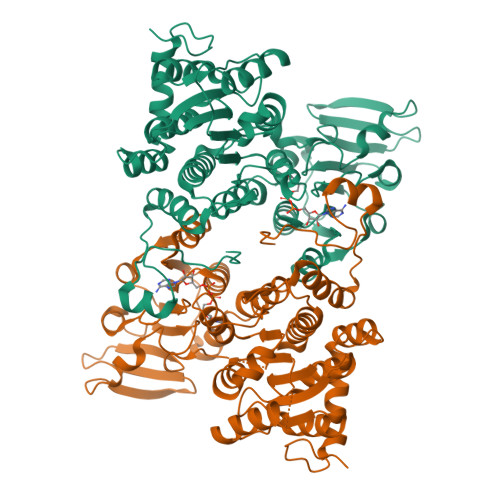
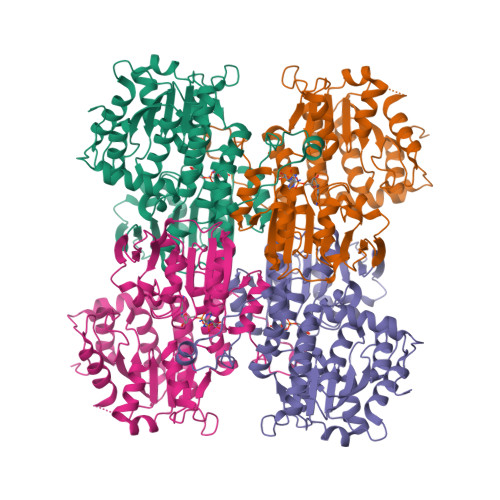
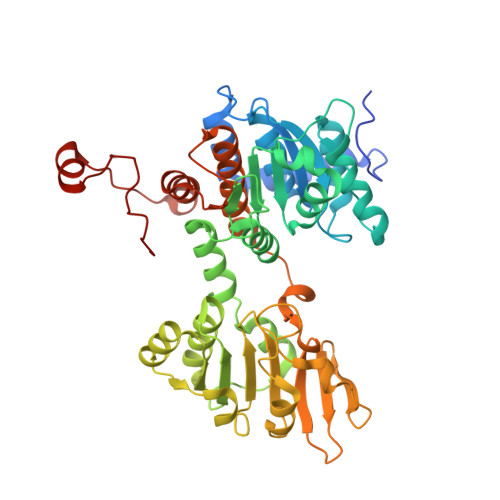
Crystal structure of human S-adenosyl homocysteine hydrolase-like 1 protein
Wisniewska, M., Siponen, M.I., Arrowsmith, C.H., Berglund, H., Bountra, C., Collins, R., Edwards, A.M., Flodin, S., Flores, A., Graslund, S., Hammarstrom, M., Johansson, I., Karlberg, T., Kotenyova, T., Moche, M., Nordlund, P., Nyman, T., Persson, C., Schutz, P., Thorsell, A.G., Tresaugues, L., van der Berg, S., Wahlberg, E., Weigelt, J., Welin, M., Schuler, H.To be published.