Structural origins of nitroxide side chain dynamics on membrane protein alpha-helical sites.
Kroncke, B.M., Horanyi, P.S., Columbus, L.(2010) Biochemistry 49: 10045-10060
- PubMed: 20964375
- DOI: https://doi.org/10.1021/bi101148w
- Primary Citation of Related Structures:
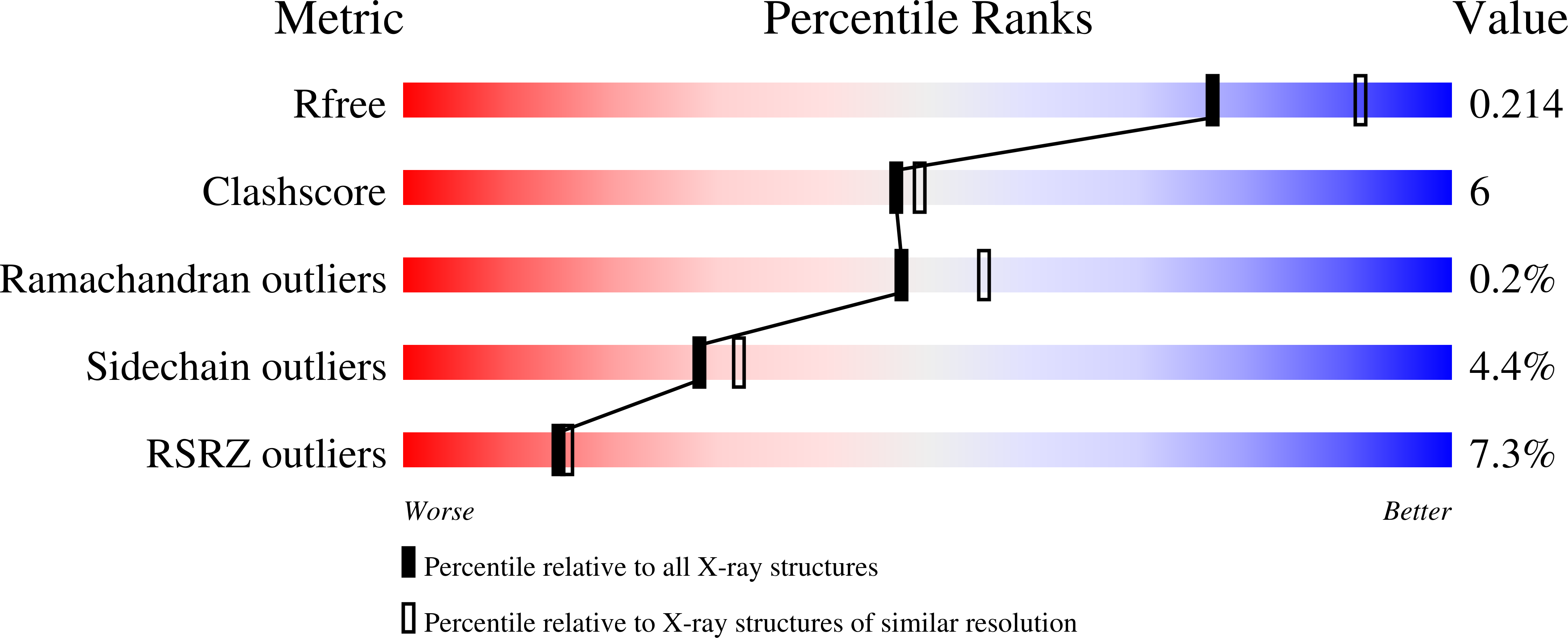
3MPN, 3MPQ - PubMed Abstract:
Understanding the structure and dynamics of membrane proteins in their native, hydrophobic environment is important to understanding how these proteins function. EPR spectroscopy in combination with site-directed spin labeling (SDSL) can measure dynamics and structure of membrane proteins in their native lipid environment; however, until now the dynamics measured have been qualitative due to limited knowledge of the nitroxide spin label's intramolecular motion in the hydrophobic environment. Although several studies have elucidated the structural origins of EPR line shapes of water-soluble proteins, EPR spectra of nitroxide spin-labeled proteins in detergents or lipids have characteristic differences from their water-soluble counterparts, suggesting significant differences in the underlying molecular motion of the spin label between the two environments. To elucidate these differences, membrane-exposed α-helical sites of the leucine transporter, LeuT, from Aquifex aeolicus, were investigated using X-ray crystallography, mutational analysis, nitroxide side chain derivatives, and spectral simulations in order to obtain a motional model of the nitroxide. For each crystal structure, the nitroxide ring of a disulfide-linked spin label side chain (R1) is resolved and makes contacts with hydrophobic residues on the protein surface. The spin label at site I204 on LeuT makes a nontraditional hydrogen bond with the ortho-hydrogen on its nearest neighbor F208, whereas the spin label at site F177 makes multiple van der Waals contacts with a hydrophobic pocket formed with an adjacent helix. These results coupled with the spectral effect of mutating the i ± 3, 4 residues suggest that the spin label has a greater affinity for its local protein environment in the low dielectric than on a water-soluble protein surface. The simulations of the EPR spectra presented here suggest the spin label oscillates about the terminal bond nearest the ring while maintaining weak contact with the protein surface. Combined, the results provide a starting point for determining a motional model for R1 on membrane proteins, allowing quantification of nitroxide dynamics in the aliphatic environment of detergent and lipids. In addition, initial contributions to a rotamer library of R1 on membrane proteins are provided, which will assist in reliably modeling the R1 conformational space for pulsed dipolar EPR and NMR paramagnetic relaxation enhancement distance determination.
Organizational Affiliation:
Department of Chemistry, University of Virginia,Charlottesville, Virginia 22904, United States.