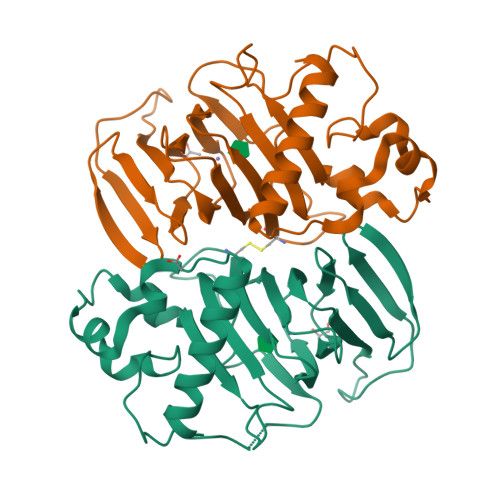
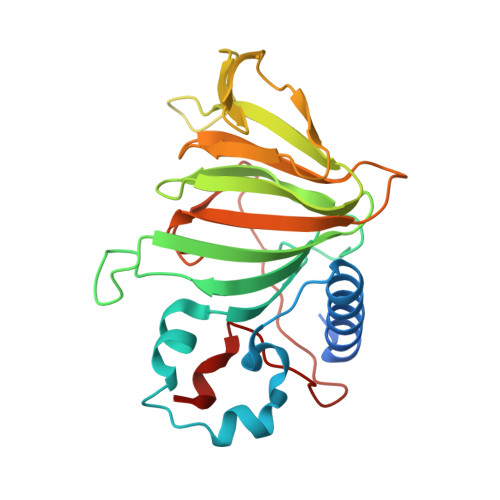
Structure-based annotation of a novel sugar isomerase from the pathogenic E. coli O157:H7.
van Staalduinen, L.M., Park, C.S., Yeom, S.J., Adams-Cioaba, M.A., Oh, D.K., Jia, Z.(2010) J Mol Biology 401: 866-881
- PubMed: 20615418
- DOI: https://doi.org/10.1016/j.jmb.2010.06.063
- Primary Citation of Related Structures:
3KMH, 3MPB - PubMed Abstract:
Prokaryotes can use a variety of sugars as carbon sources in order to provide a selective survival advantage. The gene z5688 found in the pathogenic Escherichia coli O157:H7 encodes a "hypothetical" protein of unknown function. Sequence analysis identified the gene product as a putative member of the cupin superfamily of proteins, but no other functional information was known. We have determined the crystal structure of the Z5688 protein at 1.6 A resolution and identified the protein as a novel E. coli sugar isomerase (EcSI) through overall fold analysis and secondary-structure matching. Extensive substrate screening revealed that EcSI is capable of acting on d-lyxose and d-mannose. The complex structure of EcSI with fructose allowed the identification of key active-site residues, and mutagenesis confirmed their importance. The structure of EcSI also suggested a novel mechanism for substrate binding and product release in a cupin sugar isomerase. Supplementation of a nonpathogenic E. coli strain with EcSI enabled cell growth on the rare pentose d-lyxose.
Organizational Affiliation:
Department of Biochemistry, Queen's University, Kingston, Ontario, Canada K7L3N6.