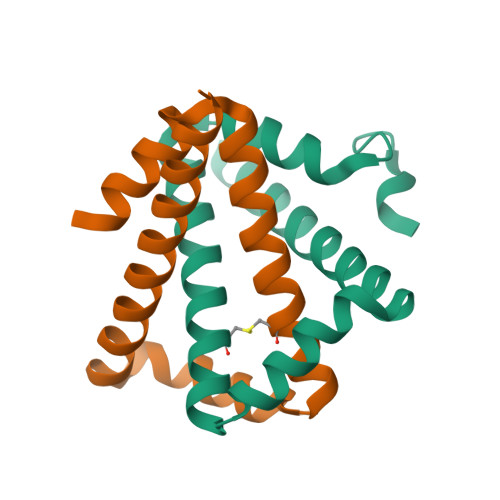
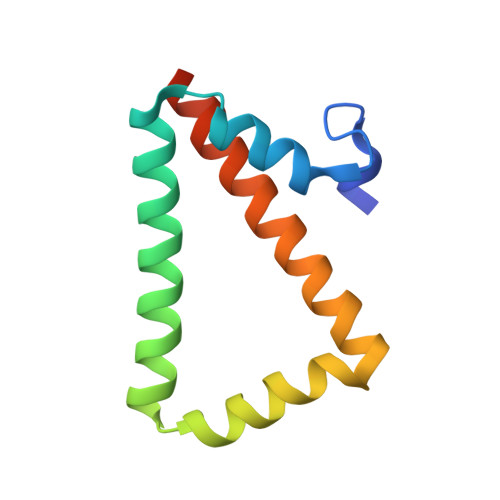
Structure and folding of a designed knotted protein.
King, N.P., Jacobitz, A.W., Sawaya, M.R., Goldschmidt, L., Yeates, T.O.(2010) Proc Natl Acad Sci U S A 107: 20732-20737
- PubMed: 21068371
- DOI: https://doi.org/10.1073/pnas.1007602107
- Primary Citation of Related Structures:
3MLG, 3MLI - PubMed Abstract:
A very small number of natural proteins have folded configurations in which the polypeptide backbone is knotted. Relatively little is known about the folding energy landscapes of such proteins, or how they have evolved. We explore those questions here by designing a unique knotted protein structure. Biophysical characterization and X-ray crystal structure determination show that the designed protein folds to the intended configuration, tying itself in a knot in the process, and that it folds reversibly. The protein folds to its native, knotted configuration approximately 20 times more slowly than a control protein, which was designed to have a similar tertiary structure but to be unknotted. Preliminary kinetic experiments suggest a complicated folding mechanism, providing opportunities for further characterization. The findings illustrate a situation where a protein is able to successfully traverse a complex folding energy landscape, though the amino acid sequence of the protein has not been subjected to evolutionary pressure for that ability. The success of the design strategy--connecting two monomers of an intertwined homodimer into a single protein chain--supports a model for evolution of knotted structures via gene duplication.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California, Los Angeles, CA 90095-1569, USA.