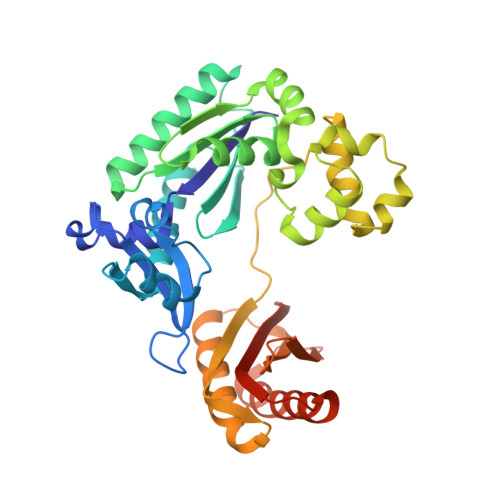
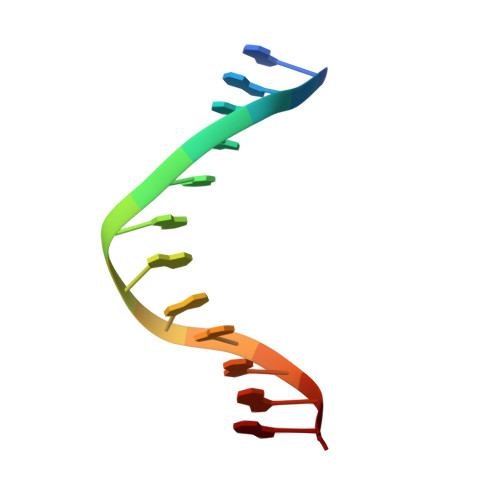
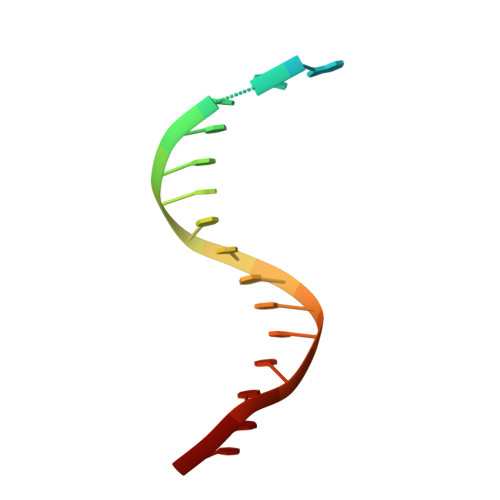
Structural insight into dynamic bypass of the major cisplatin-DNA adduct by Y-family polymerase Dpo4.
Wong, J.H., Brown, J.A., Suo, Z., Blum, P., Nohmi, T., Ling, H.(2010) EMBO J 29: 2059-2069
- PubMed: 20512114
- DOI: https://doi.org/10.1038/emboj.2010.101
- Primary Citation of Related Structures:
3M9M, 3M9N, 3M9O - PubMed Abstract:
Y-family DNA polymerases bypass Pt-GG, the cisplatin-DNA double-base lesion, contributing to the cisplatin resistance in tumour cells. To reveal the mechanism, we determined three structures of the Y-family DNA polymerase, Dpo4, in complex with Pt-GG DNA. The crystallographic snapshots show three stages of lesion bypass: the nucleotide insertions opposite the 3'G (first insertion) and 5'G (second insertion) of Pt-GG, and the primer extension beyond the lesion site. We observed a dynamic process, in which the lesion was converted from an open and angular conformation at the first insertion to a depressed and nearly parallel conformation at the subsequent reaction stages to fit into the active site of Dpo4. The DNA translocation-coupled conformational change may account for additional inhibition on the second insertion reaction. The structures illustrate that Pt-GG disturbs the replicating base pair in the active site, which reduces the catalytic efficiency and fidelity. The in vivo relevance of Dpo4-mediated Pt-GG bypass was addressed by a dpo-4 knockout strain of Sulfolobus solfataricus, which exhibits enhanced sensitivity to cisplatin and proteomic alterations consistent with genomic stress.
Organizational Affiliation:
Department of Biochemistry, University of Western Ontario, London, Ontario, Canada.