Structure of anticancer ruthenium half-sandwich complex bound to glycogen synthase kinase 3beta
Atilla-Gokcumen, G.E., Di Costanzo, L., Meggers, E.(2011) J Biol Inorg Chem 16: 45-50
- PubMed: 20821241
- DOI: https://doi.org/10.1007/s00775-010-0699-x
- Primary Citation of Related Structures:
3M1S - PubMed Abstract:
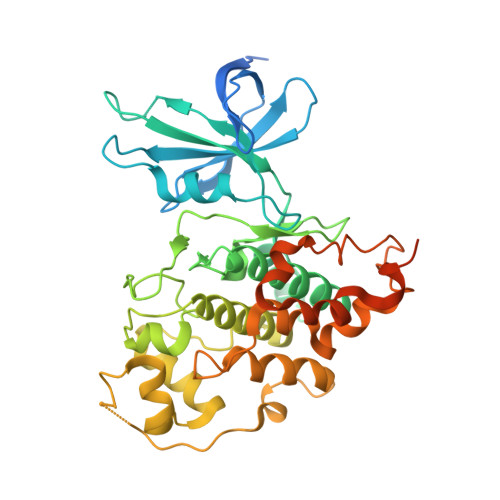
The 3.15-Å-resolution crystal structure of the R enantiomer of the highly bioactive and antiproliferative half-sandwich ruthenium complex DW12 bound to the ATP binding site of glycogen synthase kinase 3β (GSK-3β) is reported and the binding is compared with the GSK-3β binding of staurosporine and other organic inhibitors. The structure reveals a close packing of the organometallic inhibitor in the ATP binding site of GSK-3β via an induced-fit mechanism. The molecular structure of (R)-DW12 with the CO ligand oriented perpendicular to the pyridocarbazole heterocycle allows the complex to stretch the whole distance sandwiched between the faces of the N- and C-terminal lobes and to interact tightly with the flexible glycine-rich loop, which is uncommon for the interaction of GSK-3β with organic inhibitors.
Organizational Affiliation:
Department of Chemistry, University of Pennsylvania, 231 S. 34th Street, Philadelphia, PA 19104, USA.