Amyloid Fibril Recognition with the Conformational B10 Antibody Fragment Depends on Electrostatic Interactions.
Haupt, C., Morgado, I., Kumar, S.T., Parthier, C., Bereza, M., Hortschansky, P., Stubbs, M.T., Horn, U., Fandrich, M.(2010) J Mol Biol
- PubMed: 21059358
- DOI: https://doi.org/10.1016/j.jmb.2010.10.059
- Primary Citation of Related Structures:
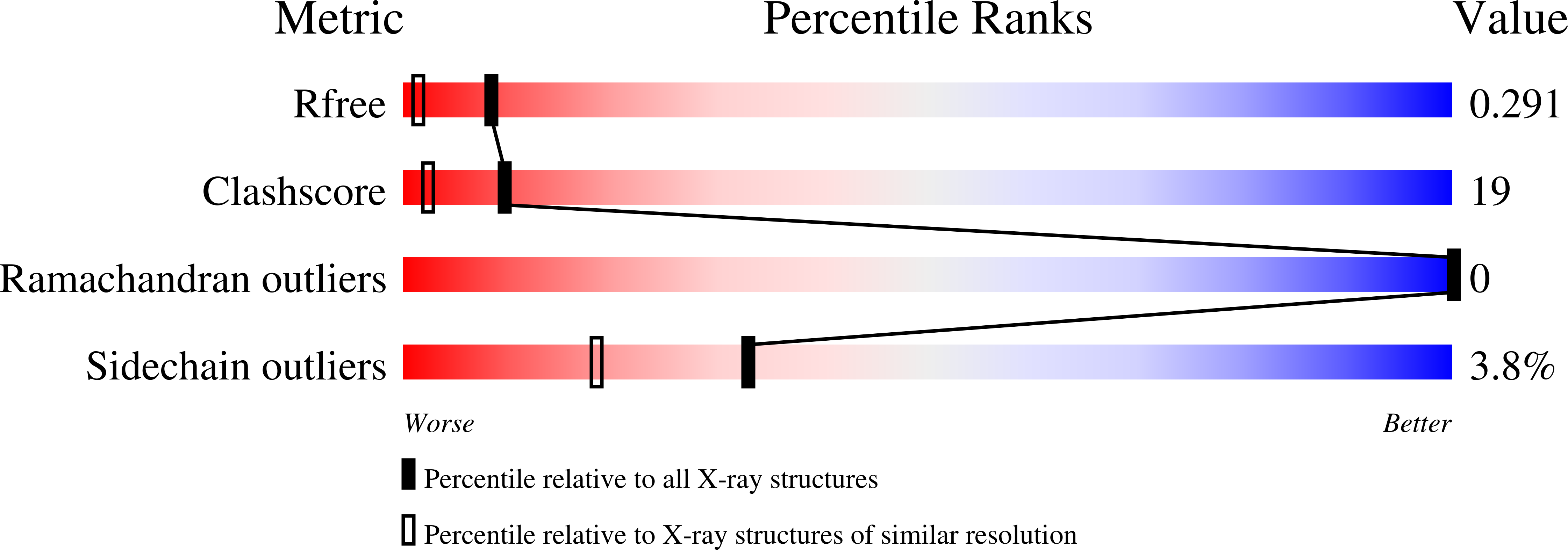
3LN9 - PubMed Abstract:
Amyloid fibrils are naturally occurring polypeptide scaffolds with considerable importance for human health and disease. These supermolecular assemblies are β-sheet rich and characterized by a high structural order. Clinical diagnosis and emerging therapeutic strategies of amyloid-dependent diseases, such as Alzheimer's, rely on the specific recognition of amyloid structures by other molecules. Recently, we generated the B10 antibody fragment, which selectively binds to Alzheimer's Aβ(1-40) amyloid fibrils but does not explicitly recognize other protein conformers, such as oligomers and disaggregated Aβ peptide. B10 presents poly-amyloid specific binding and interacts with fibrillar structures consisting of different polypeptide chains. To determine the molecular basis behind its specificity, we have analyzed the molecular properties of B10 with a battery of biochemical and biophysical techniques, ranging from X-ray crystallography to chemical modification studies. We find that fibril recognition depends on positively charged residues within the B10 antigen binding site. Mutation of these basic residues into alanine potently impairs fibril binding, and reduced B10-fibril interactions are also observed when the fibril carboxyl groups are covalently masked by a chemical modification approach. These data imply that the B10 conformational specificity for amyloid fibrils depends upon specific electrostatic interactions with an acidic moiety, which is common to different amyloid fibrils.
Organizational Affiliation:
Max-Planck Research Unit for Enzymology of Protein Folding, 06120 Halle, Germany.