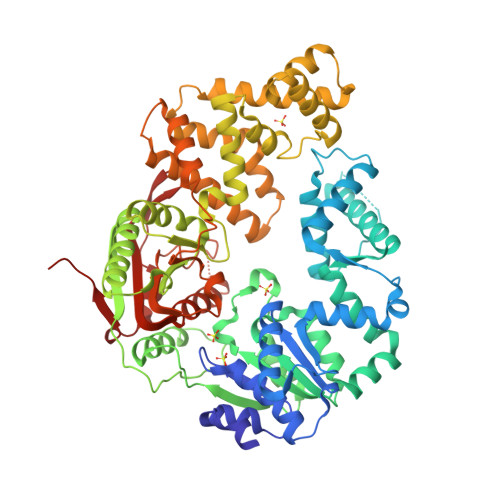
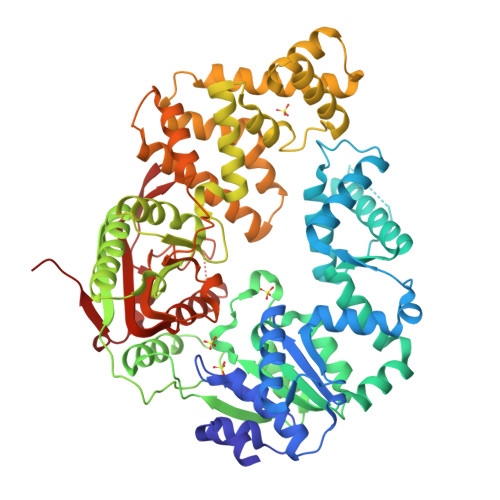
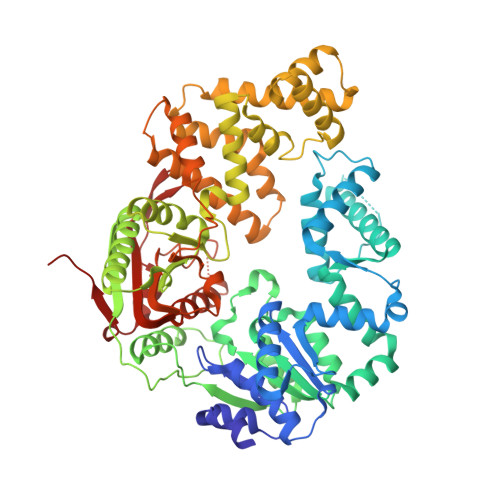
Rotations of the 2B sub-domain of E. coli UvrD helicase/translocase coupled to nucleotide and DNA binding.
Jia, H., Korolev, S., Niedziela-Majka, A., Maluf, N.K., Gauss, G.H., Myong, S., Ha, T., Waksman, G., Lohman, T.M.(2011) J Mol Biology 411: 633-648
- PubMed: 21704638
- DOI: https://doi.org/10.1016/j.jmb.2011.06.019
- Primary Citation of Related Structures:
3LFU - PubMed Abstract:
Escherichia coli UvrD is a superfamily 1 DNA helicase and single-stranded DNA (ssDNA) translocase that functions in DNA repair and plasmid replication and as an anti-recombinase by removing RecA protein from ssDNA. UvrD couples ATP binding and hydrolysis to unwind double-stranded DNA and translocate along ssDNA with 3'-to-5' directionality. Although a UvrD monomer is able to translocate along ssDNA rapidly and processively, DNA helicase activity in vitro requires a minimum of a UvrD dimer. Previous crystal structures of UvrD bound to a ssDNA/duplex DNA junction show that its 2B sub-domain exists in a "closed" state and interacts with the duplex DNA. Here, we report a crystal structure of an apo form of UvrD in which the 2B sub-domain is in an "open" state that differs by an ∼160° rotation of the 2B sub-domain. To study the rotational conformational states of the 2B sub-domain in various ligation states, we constructed a series of double-cysteine UvrD mutants and labeled them with fluorophores such that rotation of the 2B sub-domain results in changes in fluorescence resonance energy transfer. These studies show that the open and closed forms can interconvert in solution, with low salt favoring the closed conformation and high salt favoring the open conformation in the absence of DNA. Binding of UvrD to DNA and ATP binding and hydrolysis also affect the rotational conformational state of the 2B sub-domain, suggesting that 2B sub-domain rotation is coupled to the function of this nucleic acid motor enzyme.
Organizational Affiliation:
Department of Biochemistry and Molecular Biophysics, Washington University School of Medicine, 660 South Euclid Avenue, St. Louis, MO 63110, USA.