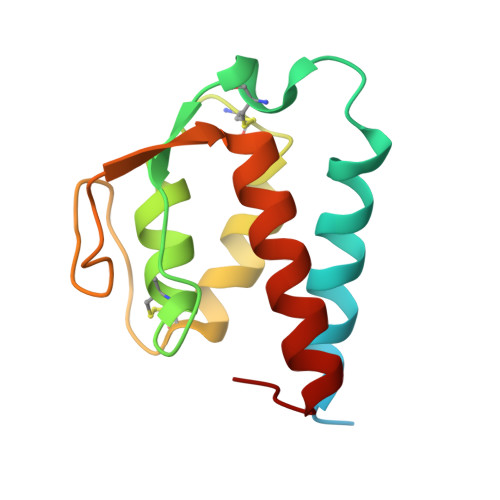
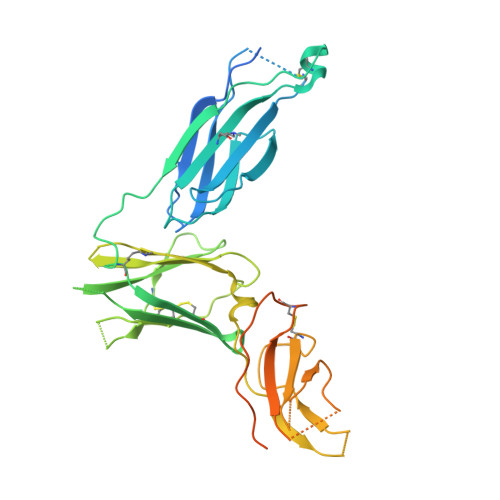
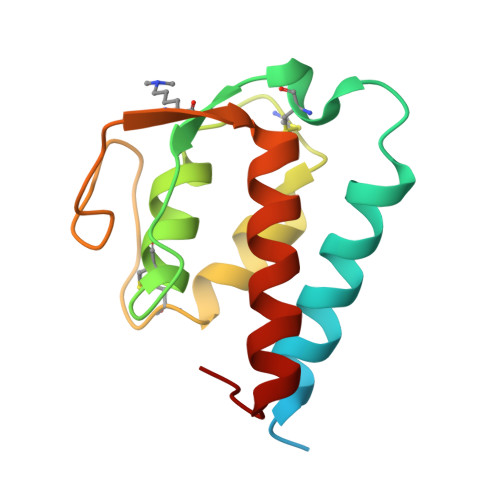
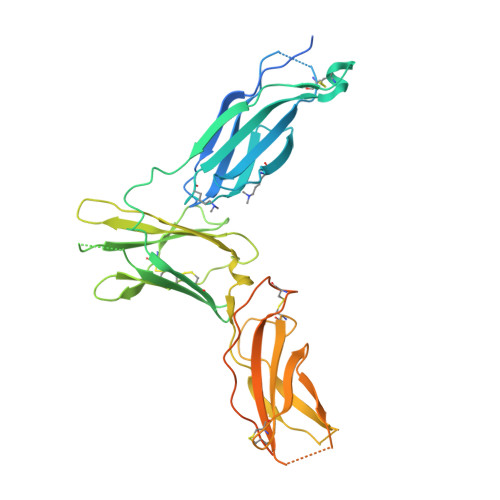
Molecular basis for shared cytokine recognition revealed in the structure of an unusually high affinity complex between IL-13 and IL-13Ralpha2.
Lupardus, P.J., Birnbaum, M.E., Garcia, K.C.(2010) Structure 18: 332-342
- PubMed: 20223216
- DOI: https://doi.org/10.1016/j.str.2010.01.003
- Primary Citation of Related Structures:
3LB6 - PubMed Abstract:
Interleukin-13 is a cytokine important for development of T helper cell type 2 (Th2) responses and plays a critical role in asthma and allergy. The IL-13 Receptor alpha2 (IL-13Ralpha2) is a receptor for IL-13 lacking canonical Jak/STAT signaling functions. Here we present the crystal structure along with a mutational and biophysical analysis of the IL-13/IL-13Ralpha2 complex. While retaining a similar mode of IL-13 binding to its related signaling receptor, IL-13Ralpha1, IL-13Ralpha2 uses peripheral receptor residues unused in the IL-13/IL-13Ralpha1 complex to generate a larger and more complementary interface for IL-13. This results in a four orders of magnitude increase in affinity, to the femtomolar level, compared to IL-13Ralpha1. Alanine scanning mutagenesis of the IL-13 interface reveals several common "hotspot" residues important for binding to both receptors, but also identifies a prominent IL-13Ralpha2-specific contact. These results provide a framework for development of receptor subtype-selective IL-13 antagonists and indicate a decoy function for IL-13Ralpha2.
- Howard Hughes Medical Institute, Department of Molecular and Cellular Physiology, Department of Structural Biology, and Program in Immunology, Stanford University School of Medicine, Stanford, CA 94305, USA.
Organizational Affiliation: