Structure of grouper iridovirus purine nucleoside phosphorylase
Kang, Y.N., Zhang, Y., Allan, P.W., Parker, W.B., Ting, J.W., Chang, C.Y., Ealick, S.E.(2010) Acta Crystallogr D Biol Crystallogr 66: 155-162
- PubMed: 20124695
- DOI: https://doi.org/10.1107/S0907444909048276
- Primary Citation of Related Structures:
3KHS - PubMed Abstract:
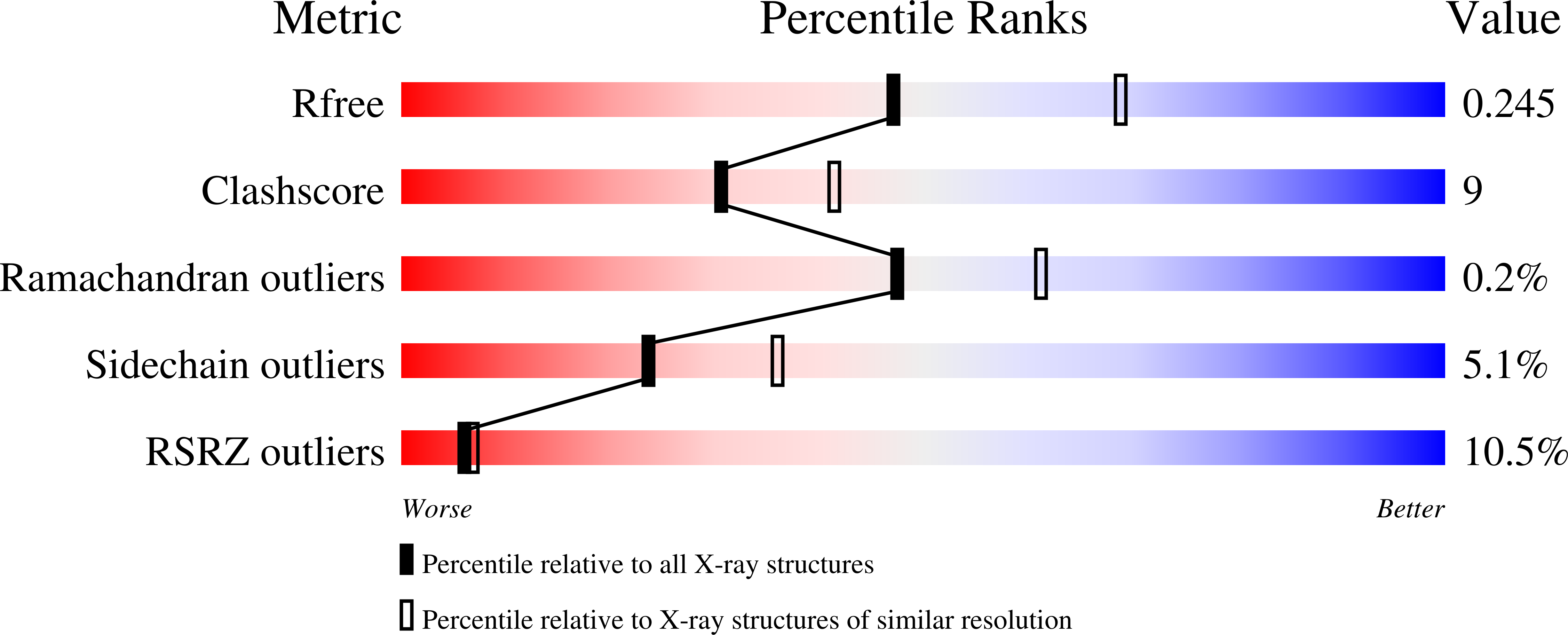
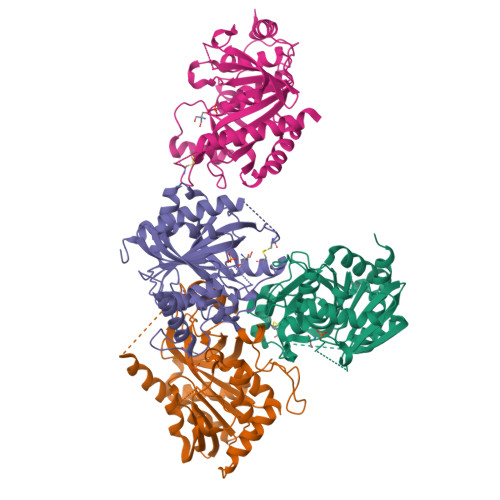
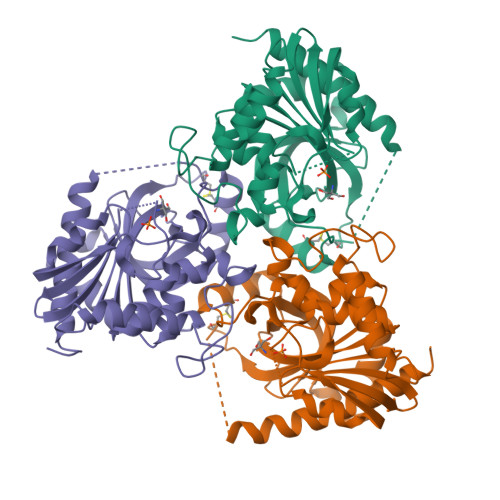
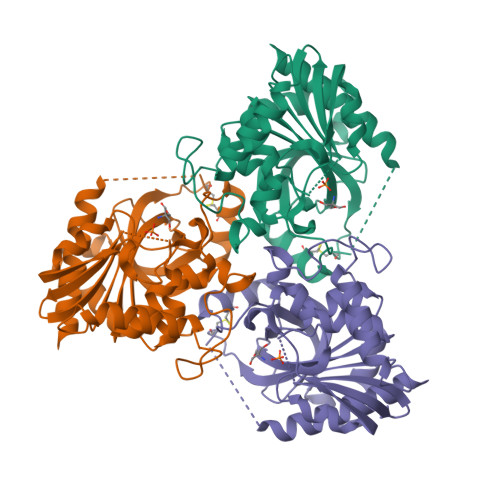
Purine nucleoside phosphorylase (PNP) catalyzes the reversible phosphorolysis of purine ribonucleosides to the corresponding free bases and ribose 1-phosphate. The crystal structure of grouper iridovirus PNP (givPNP), corresponding to the first PNP gene to be found in a virus, was determined at 2.4 A resolution. The crystals belonged to space group R3, with unit-cell parameters a = 193.0, c = 105.6 A, and contained four protomers per asymmetric unit. The overall structure of givPNP shows high similarity to mammalian PNPs, having an alpha/beta structure with a nine-stranded mixed beta-barrel flanked by a total of nine alpha-helices. The predicted phosphate-binding and ribose-binding sites are occupied by a phosphate ion and a Tris molecule, respectively. The geometrical arrangement and hydrogen-bonding patterns of the phosphate-binding site are similar to those found in the human and bovine PNP structures. The enzymatic activity assay of givPNP on various substrates revealed that givPNP can only accept 6-oxopurine nucleosides as substrates, which is also suggested by its amino-acid composition and active-site architecture. All these results suggest that givPNP is a homologue of mammalian PNPs in terms of amino-acid sequence, molecular mass, substrate specificity and overall structure, as well as in the composition of the active site.
Organizational Affiliation:
Department of Chemistry and Chemical Biology, Cornell University, Ithaca, NY 14853-1301, USA.