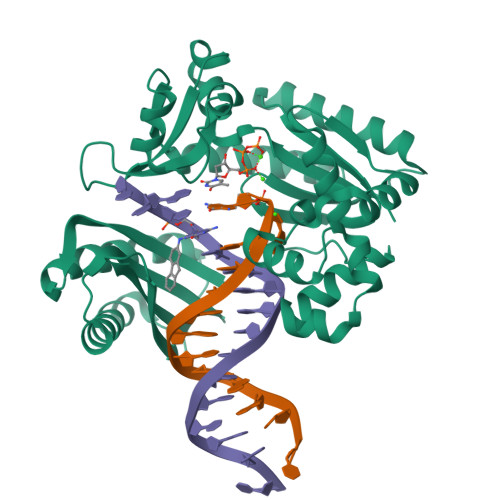
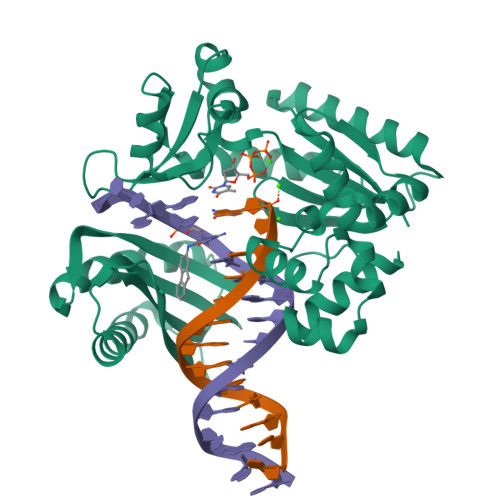
Mechanism of error-free and semitargeted mutagenic bypass of an aromatic amine lesion by Y-family polymerase Dpo4.
Rechkoblit, O., Kolbanovskiy, A., Malinina, L., Geacintov, N.E., Broyde, S., Patel, D.J.(2010) Nat Struct Mol Biol 17: 379-388
- PubMed: 20154704
- DOI: https://doi.org/10.1038/nsmb.1771
- Primary Citation of Related Structures:
3KHG, 3KHH, 3KHL, 3KHR - PubMed Abstract:
The aromatic amine carcinogen 2-aminofluorene (AF) forms covalent adducts with DNA, predominantly with guanine at the C8 position. Such lesions are bypassed by Y-family polymerases such as Dpo4 via error-free and error-prone mechanisms. We show that Dpo4 catalyzes elongation from a correct 3'-terminal cytosine opposite [AF]G in a nonrepetitive template sequence with low efficiency. This extension leads to cognate full-length product, as well as mis-elongated products containing base mutations and deletions. Crystal structures of the Dpo4 ternary complex, with the 3'-terminal primer cytosine base opposite [AF]G in the anti conformation and with the AF moiety positioned in the major groove, reveal both accurate and misalignment-mediated mutagenic extension pathways. The mutagenic template-primer-dNTP arrangement is promoted by interactions between the polymerase and the bulky lesion rather than by a base pair-stabilized misaligment. Further extension leads to semitargeted mutations via this proposed polymerase-guided mechanism.
Organizational Affiliation:
Structural Biology Program, Memorial Sloan-Kettering Cancer Center, New York, New York, USA.