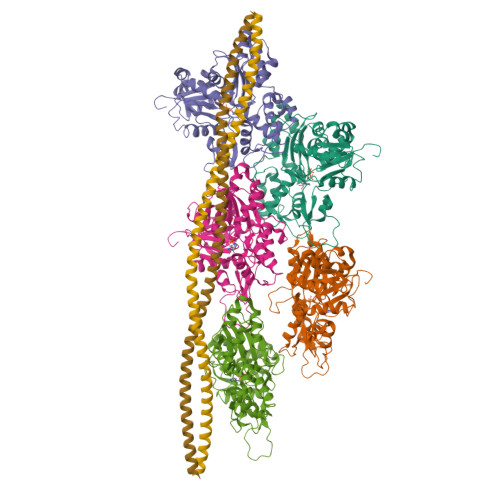
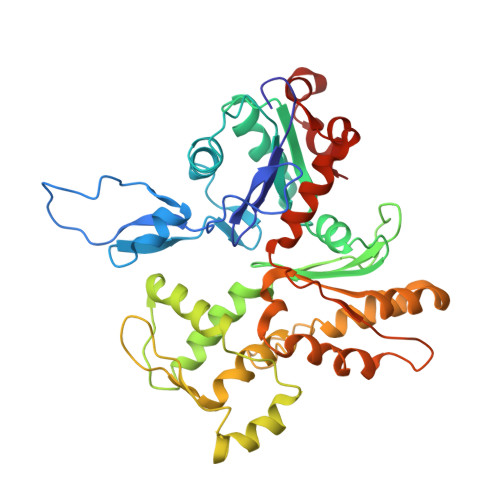
Cryo-EM Structures of the Actin:Tropomyosin Filament Reveal the Mechanism for the Transition from C- to M-State.
Sousa, D.R., Stagg, S.M., Stroupe, M.E.(2013) J Mol Biology 425: 4544-4555
- PubMed: 24021812
- DOI: https://doi.org/10.1016/j.jmb.2013.08.020
- Primary Citation of Related Structures:
3J4K - PubMed Abstract:
Tropomyosin (Tm) is a key factor in the molecular mechanisms that regulate the binding of myosin motors to actin filaments (F-Actins) in most eukaryotic cells. This regulation is achieved by the azimuthal repositioning of Tm along the actin (Ac):Tm:troponin (Tn) thin filament to block or expose myosin binding sites on Ac. In striated muscle, including involuntary cardiac muscle, Tm regulates muscle contraction by coupling Ca(2+) binding to Tn with myosin binding to the thin filament. In smooth muscle, the switch is the posttranslational modification of the myosin. Depending on the activation state of Tn and the binding state of myosin, Tm can occupy the blocked, closed, or open position on Ac. Using native cryogenic 3DEM (three-dimensional electron microscopy), we have directly resolved and visualized cardiac and gizzard muscle Tm on filamentous Ac in the position that corresponds to the closed state. From the 8-Å-resolution structure of the reconstituted Ac:Tm filament formed with gizzard-derived Tm, we discuss two possible mechanisms for the transition from closed to open state and describe the role Tm plays in blocking myosin tight binding in the closed-state position.
Organizational Affiliation:
Department of Biological Science and Institute of Molecular Biophysics, Florida State University, 91 Chieftan Way, Tallahassee, FL 32306, USA; Department of Physiology and Biophysics, Boston University School of Medicine, 72 East Concord Street, Boston, MA 02118-2526, USA.