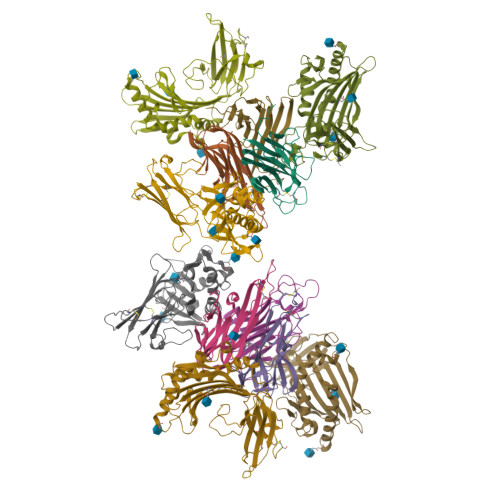
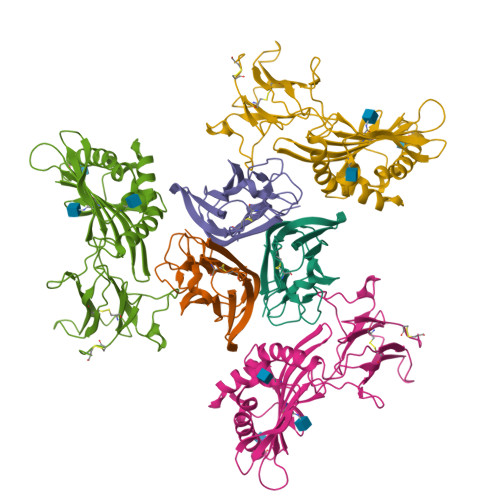
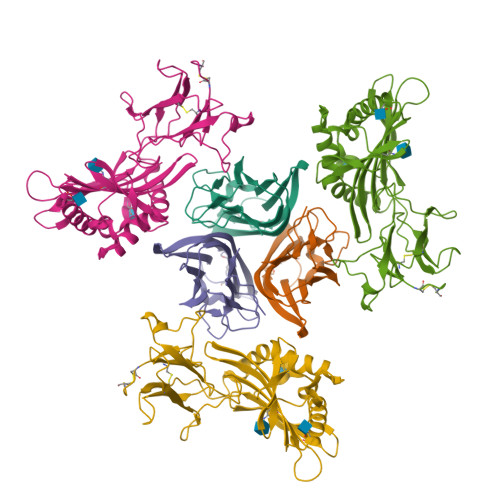
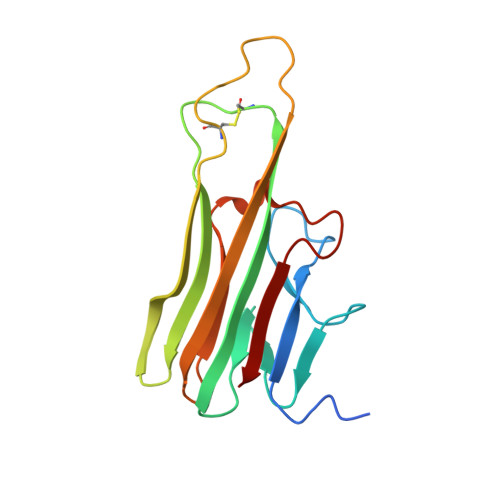
Crystal structure of TNFalpha complexed with a poxvirus MHC-related TNF binding protein
Yang, Z., West, A.P., Bjorkman, P.J.(2009) Nat Struct Mol Biol 16: 1189-1191
- PubMed: 19838188
- DOI: https://doi.org/10.1038/nsmb.1683
- Primary Citation of Related Structures:
3IT8 - PubMed Abstract:
The poxvirus 2L protein binds tumor necrosis factor-alpha (TNFalpha) to inhibit host antiviral and immune responses. The 2.8-A 2L-TNFalpha structure reveals three symmetrically arranged 2L molecules per TNFalpha trimer. 2L resembles class I major histocompatibility complex (MHC) molecules but lacks a peptide-binding groove and beta2-microglobulin light chain. Overlap between the 2L and host TNF receptor-binding sites on TNFalpha rationalizes 2L inhibition of TNFalpha-TNF receptor interactions and prevention of TNFalpha-induced immune responses.
Organizational Affiliation:
Division of Biology 114-96, California Institute of Technology, Pasadena, California, USA.