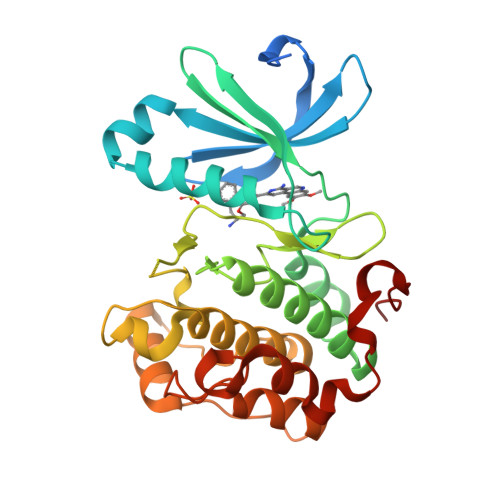
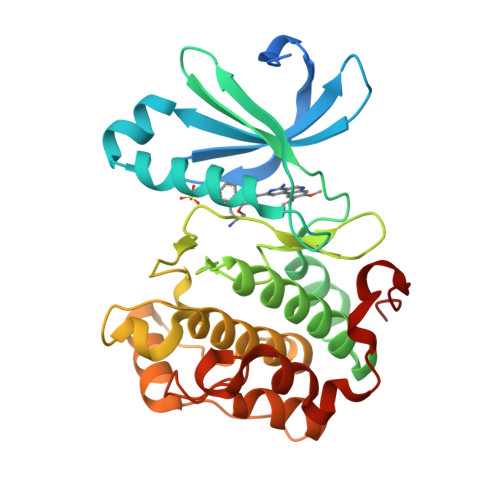
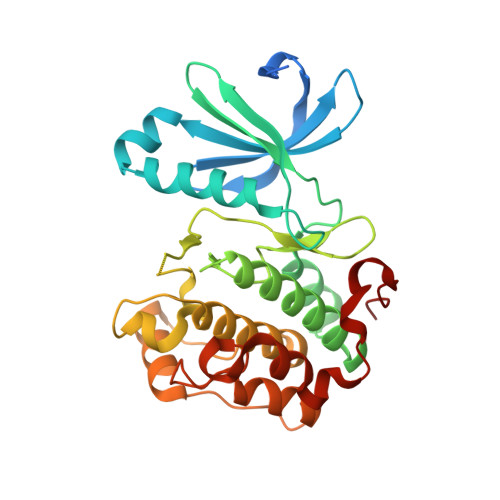
The identification of 8,9-dimethoxy-5-(2-aminoalkoxy-pyridin-3-yl)-benzo[c][2,7]naphthyridin-4-ylamines as potent inhibitors of 3-phosphoinositide-dependent kinase-1 (PDK-1).
Nittoli, T., Dushin, R.G., Ingalls, C., Cheung, K., Floyd, M.B., Fraser, H., Olland, A., Hu, Y., Grosu, G., Han, X., Arndt, K., Guo, B., Wissner, A.(2010) Eur J Med Chem 45: 1379-1386
- PubMed: 20074837
- DOI: https://doi.org/10.1016/j.ejmech.2009.12.036
- Primary Citation of Related Structures:
3ION, 3IOP - PubMed Abstract:
A series of 8,9-dimethoxy-5-(2-aminoalkoxy-pyridin-3-yl)-benzo[c][2,7]naphthyridin-4-ylamine-based inhibitors of 3-phosphoinositide-dependent kinase-1 (PDK-1) has been identified. Several examples appear to be potent and relatively selective inhibitors of PDK-1 over the related AGC kinases PKA, PKB/AKT, and p70S6K. The introduction of a stereochemical center beside the amino substituent on the aminoalkoxy-side chain had little effect upon the inhibitory activity against these enzymes, and X-ray crystallographic analyses of a representative pair of enantiomeric inhibitors bound to the active site of PDK-1 revealed comparable binding modes for each enantiomer.
Organizational Affiliation:
Chemical Sciences, Wyeth Research, 401 N. Middletown Road, Pearl River, NY 10965, USA. nittoli@hotmail.com