The crystal structure of dihydrodipicolinate reductase from the human-pathogenic bacterium Bartonella henselae strain Houston-1 at 2.3 angstrom resolution.
Cala, A.R., Nadeau, M.T., Abendroth, J., Staker, B.L., Reers, A.R., Weatherhead, A.W., Dobson, R.C., Myler, P.J., Hudson, A.O.(2016) Acta Crystallogr F Struct Biol Commun 72: 885-891
- PubMed: 27917836
- DOI: https://doi.org/10.1107/S2053230X16018525
- Primary Citation of Related Structures:
3IJP - PubMed Abstract:
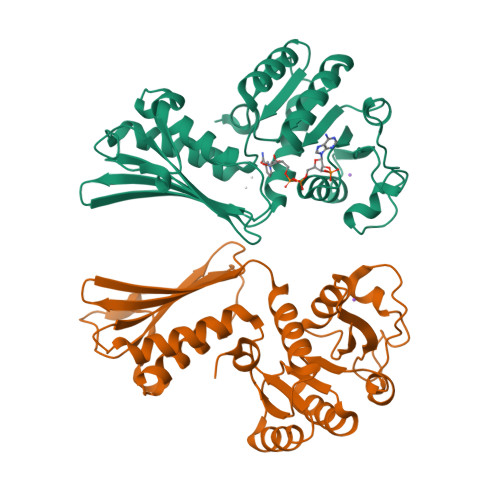
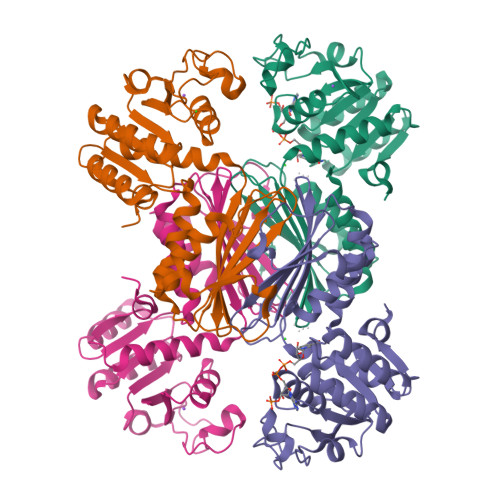
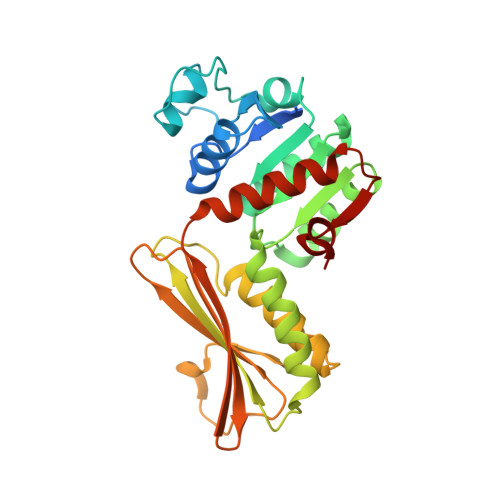
In bacteria, the second committed step in the diaminopimelate/lysine anabolic pathways is catalyzed by the enzyme dihydrodipicolinate reductase (DapB). DapB catalyzes the reduction of dihydrodipicolinate to yield tetrahydrodipicolinate. Here, the cloning, expression, purification, crystallization and X-ray diffraction analysis of DapB from the human-pathogenic bacterium Bartonella henselae, the causative bacterium of cat-scratch disease, are reported. Protein crystals were grown in conditions consisting of 5%(w/v) PEG 4000, 200 mM sodium acetate, 100 mM sodium citrate tribasic pH 5.5 and were shown to diffract to ∼2.3 Å resolution. They belonged to space group P4 3 22, with unit-cell parameters a = 109.38, b = 109.38, c = 176.95 Å. R r.i.m. was 0.11, R work was 0.177 and R free was 0.208. The three-dimensional structural features of the enzymes show that DapB from B. henselae is a tetramer consisting of four identical polypeptides. In addition, the substrate NADP + was found to be bound to one monomer, which resulted in a closed conformational change in the N-terminal domain.
Organizational Affiliation:
Thomas H. Gosnell School of Life Sciences, Rochester Institute of Technology, 85 Lomb Memorial Drive, Rochester, NY 14623-5603, USA.