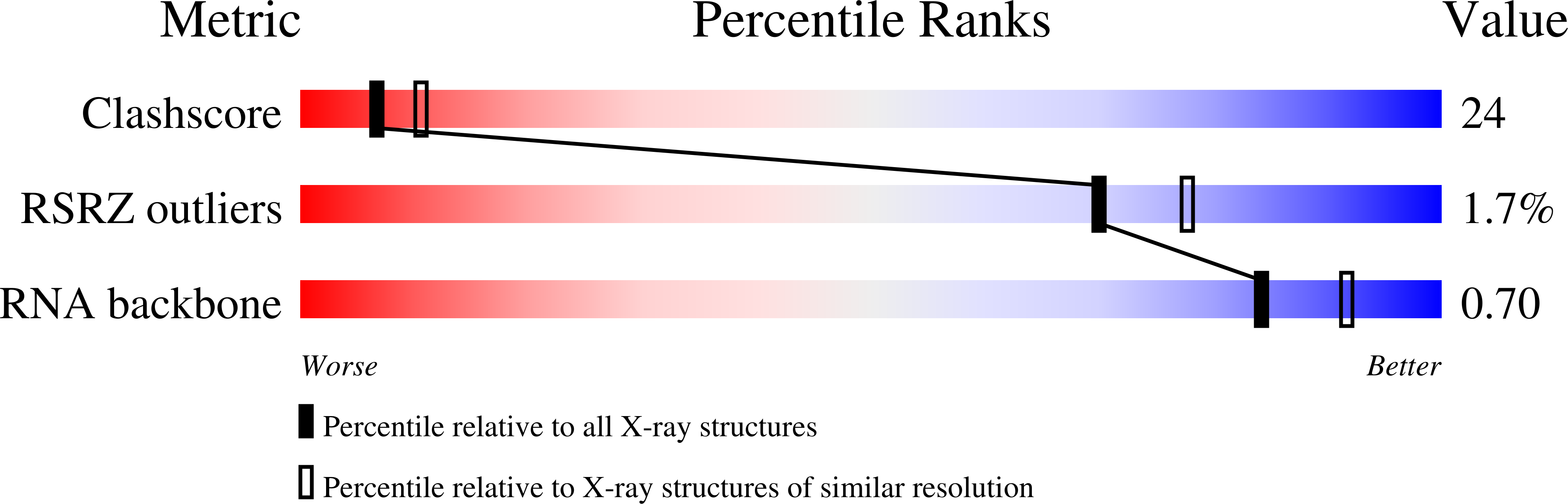Single-atom imino substitutions at A9 and A10 reveal distinct effects on the fold and function of the hairpin ribozyme catalytic core.
Spitale, R.C., Volpini, R., Mungillo, M.V., Krucinska, J., Cristalli, G., Wedekind, J.E.(2009) Biochemistry 48: 7777-7779
- PubMed: 19634899
- DOI: https://doi.org/10.1021/bi9011622
- Primary Citation of Related Structures:
3I2Q, 3I2R, 3I2S, 3I2U - PubMed Abstract:
The hairpin ribozyme cleaves a phosphodiester bond within a cognate substrate. Structural and biochemical data indicate the conserved A9 and A10 bases reside close to the scissile bond but make distinct contributions to catalysis. To investigate these residues, we replaced the imino moiety of each base with N1-deazaadenosine. This single-atom change resulted in an 8-fold loss in k(obs) for A9 and displacement of the base from the active site; no effects were observed for A10. We propose that the imino moiety of A9 promotes a key water-mediated contact that favors transition-state formation, which suggests an enhanced chemical repertoire for RNA.
Organizational Affiliation:
Department of Biochemistry & Biophysics, 601 Elmwood Avenue, Box 712, Rochester New York 14642, USA.