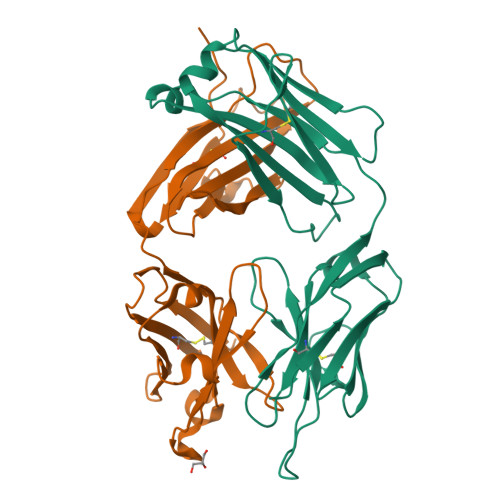
Noncanonical conformation of CDR L1 in the anti-IL-23 antibody CNTO4088.
Teplyakov, A., Obmolova, G., Rogers, A., Gilliland, G.L.(2010) Acta Crystallogr Sect F Struct Biol Cryst Commun 66: 229-232
- PubMed: 20208149
- DOI: https://doi.org/10.1107/S1744309109054141
- Primary Citation of Related Structures:
3I2C - PubMed Abstract:
CNTO4088 is a monoclonal antibody to human IL-23. The X-ray structure of the Fab fragment revealed an unusual noncanonical conformation of CDR L1. Most antibodies with the kappa light chain exhibit a canonical structure for CDR L1 in which residue 29 anchors the CDR loop to the framework. Analysis of the residues believed to define the conformation of CDR L1 did not explain why it should not adopt a canonical conformation in this antibody. This makes CNTO4088 a benchmark case for developing prediction methods and structure-modeling tools.
Organizational Affiliation:
Centocor R&D Inc., 145 King of Prussia Road, Radnor, PA 19087, USA. ateplyak@its.jnj.com