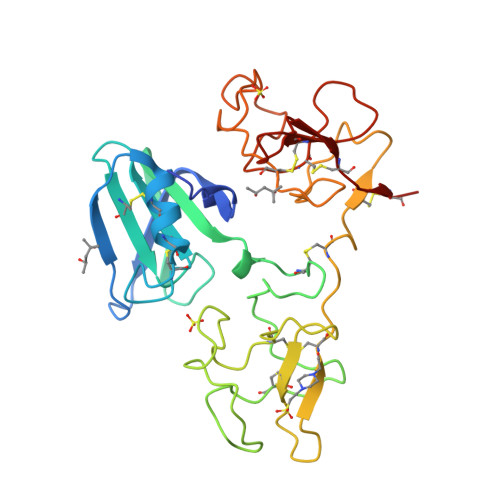
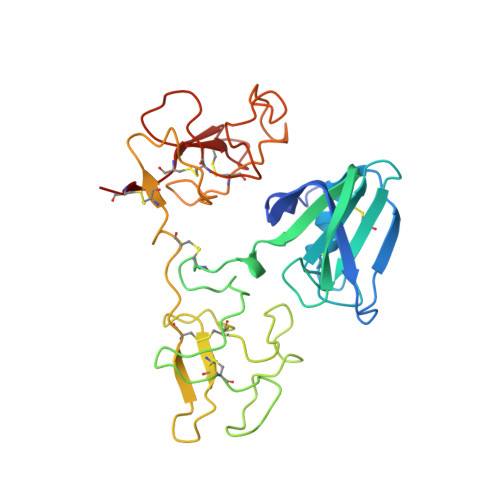
Structural basis for agonism and antagonism of hepatocyte growth factor.
Tolbert, W.D., Daugherty-Holtrop, J., Gherardi, E., Vande Woude, G., Xu, H.E.(2010) Proc Natl Acad Sci U S A 107: 13264-13269
- PubMed: 20624990
- DOI: https://doi.org/10.1073/pnas.1005183107
- Primary Citation of Related Structures:
3HMR, 3HMS, 3HMT, 3HN4 - PubMed Abstract:
Hepatocyte growth factor (HGF) is an activating ligand of the Met receptor tyrosine kinase, whose activity is essential for normal tissue development and organ regeneration but abnormal activation of Met has been implicated in growth, invasion, and metastasis of many types of solid tumors. HGF has two natural splice variants, NK1 and NK2, which contain the N-terminal domain (N) and the first kringle (K1) or the first two kringle domains of HGF. NK1, which is a Met agonist, forms a head-to-tail dimer complex in crystal structures and mutations in the NK1 dimer interface convert NK1 to a Met antagonist. In contrast, NK2 is a Met antagonist, capable of inhibiting HGF's activity in cell proliferation without clear mechanism. Here we report the crystal structure of NK2, which forms a "closed" monomeric conformation through interdomain interactions between the N- domain and the second kringle domain (K2). Mutations that were designed to open up the NK2 closed conformation by disrupting the N/K2 interface convert NK2 from a Met antagonist to an agonist. Remarkably, this mutated NK2 agonist can be converted back to an antagonist by a mutation that disrupts the NK1/NK1 dimer interface. These results reveal the molecular determinants that regulate the agonist/antagonist properties of HGF NK2 and provide critical insights into the dimerization mechanism that regulates the Met receptor activation by HGF.
Organizational Affiliation:
Laboratory of Structural Sciences, Van Andel Research Institute, 333 Bostwick Avenue, Grand Rapids, MI 49503, USA.