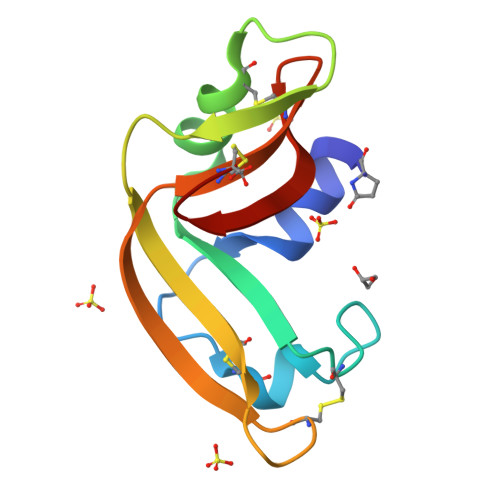
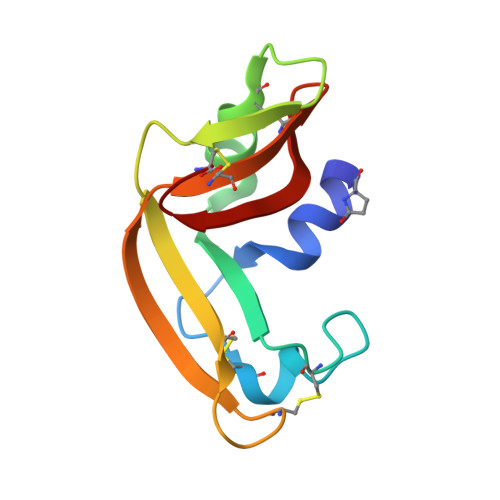
Three-state thermal unfolding of onconase.
Casares-Atienza, S., Weininger, U., Camara-Artigas, A., Balbach, J., Garcia-Mira, M.M.(2011) Biophys Chem 159: 267-274
- PubMed: 21840114
- DOI: https://doi.org/10.1016/j.bpc.2011.07.005
- Primary Citation of Related Structures:
3HG6 - PubMed Abstract:
Onconase is a member of the ribonuclease A superfamily currently in phase IIIb clinical trials as a treatment for malign mesothelioma due to its cytotoxic activity selective against tumor-cells. In this work, we have studied the equilibrium thermal unfolding of onconase using a combination of several structural and biophysical techniques. Our results indicate that at least one significantly populated intermediate, which implies the exposure of hydrophobic surface and significant changes in the environment around Trp3, occurs during the equilibrium unfolding process of this protein. The intermediate begins to populate at about 30° below the global unfolding temperature, reaching a maximum population of nearly 60%, 10° below the global unfolding temperature.
Organizational Affiliation:
Departamento de Química Física, Facultad de Ciencias, Universidad de Granada. Avda. Fuentenueva s/n. 18071 Granada, Spain.