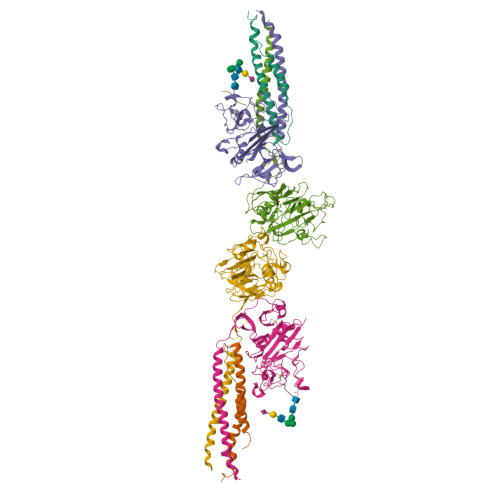
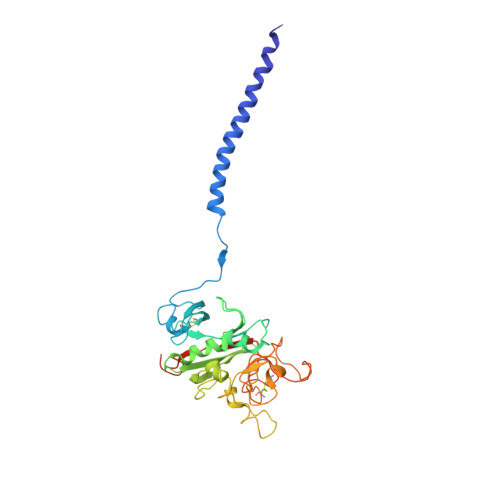
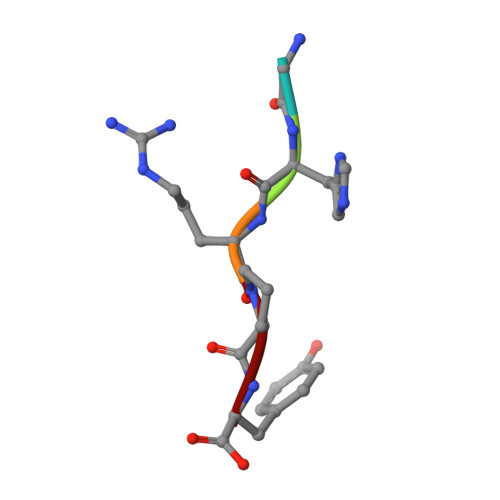
Two families of synthetic peptides that enhance fibrin turbidity and delay fibrinolysis by different mechanisms.
Pandi, L., Kollman, J.M., Lopez-Lira, F., Burrows, J.M., Riley, M., Doolittle, R.F.(2009) Biochemistry 48: 7201-7208
- PubMed: 19588915
- DOI: https://doi.org/10.1021/bi900647g
- Primary Citation of Related Structures:
3H32 - PubMed Abstract:
When fibrin clots are formed in vitro in the presence of certain positively charged peptides, the turbidity is enhanced and fibrinolysis is delayed. Here we show that these two phenomena are not always linked and that different families of peptides bring about the delay of lysis in different ways. In the case of intrinsically adhesive peptides corresponding to certain regions of the fibrinogen gammaC and betaC domains, even though these peptides bind to fibrin(ogen) and enhance turbidity, the delay in lysis is mainly due to direct inhibition of plasminogen activation. In contrast, for certain pentapeptides patterned on fibrin B knobs, the delay in lysis is a consequence of how fibrin units assemble. On their own, these B knob surrogates can induce the gelation of fibrinogen molecules. The likely cause of enhanced clot turbidity and delay in fibrinolysis was revealed by a crystal structure of the D-dimer from human fibrinogen cocrystallized with GHRPYam, the packing of which showed the direct involvement of the ligand tyrosines in antiparallel betaC-betaC interactions.
Organizational Affiliation:
Department of Chemistry and Biochemistry and Division of Biology, University of California at San Diego, La Jolla, California 92093-0314, USA.