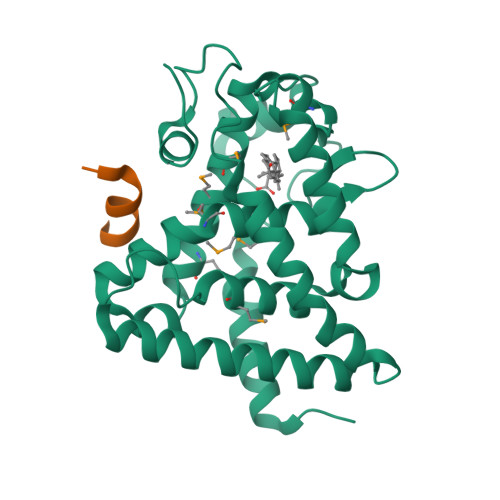
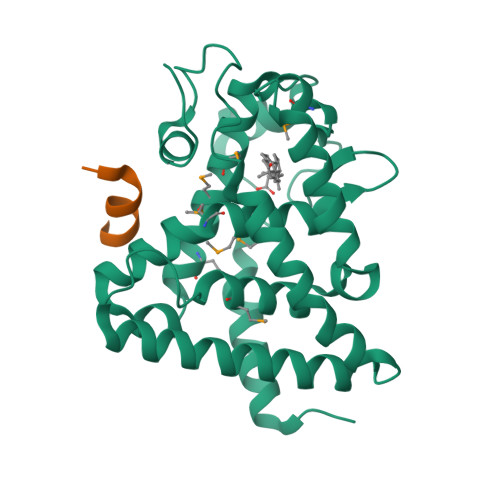
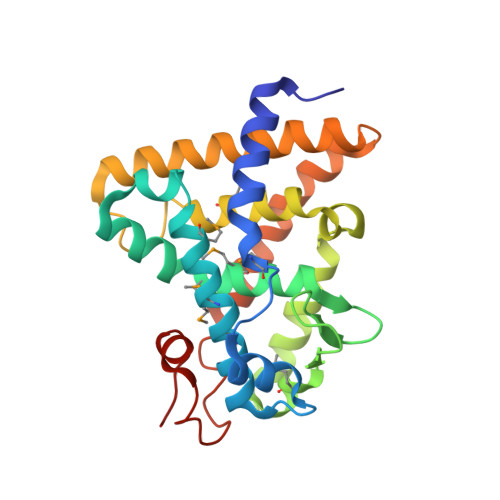
Identification of the nuclear receptor DAF-12 as a therapeutic target in parasitic nematodes.
Wang, Z., Zhou, X.E., Motola, D.L., Gao, X., Suino-Powell, K., Conneely, A., Ogata, C., Sharma, K.K., Auchus, R.J., Lok, J.B., Hawdon, J.M., Kliewer, S.A., Xu, H.E., Mangelsdorf, D.J.(2009) Proc Natl Acad Sci U S A 106: 9138-9143
- PubMed: 19497877
- DOI: https://doi.org/10.1073/pnas.0904064106
- Primary Citation of Related Structures:
3GYT, 3GYU - PubMed Abstract:
Nematode parasitism is a worldwide health problem resulting in malnutrition and morbidity in over 1 billion people. The molecular mechanisms governing infection are poorly understood. Here, we report that an evolutionarily conserved nuclear hormone receptor signaling pathway governs development of the stage 3 infective larvae (iL3) in several nematode parasites, including Strongyloides stercoralis, Ancylostoma spp., and Necator americanus. As in the free-living Caenorhabditis elegans, steroid hormone-like dafachronic acids induced recovery of the dauer-like iL3 in parasitic nematodes by activating orthologs of the nuclear receptor DAF-12. Moreover, administration of dafachronic acid markedly reduced the pathogenic iL3 population in S. stercoralis, indicating the potential use of DAF-12 ligands to treat disseminated strongyloidiasis. To understand the pharmacology of targeting DAF-12, we solved the 3-dimensional structure of the S. stercoralis DAF-12 ligand-binding domain cocrystallized with dafachronic acids. These results reveal the molecular basis for DAF-12 ligand binding and identify nuclear receptors as unique therapeutic targets in parasitic nematodes.
Organizational Affiliation:
Department of Pharmacology and Howard Hughes Medical Institute, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA.