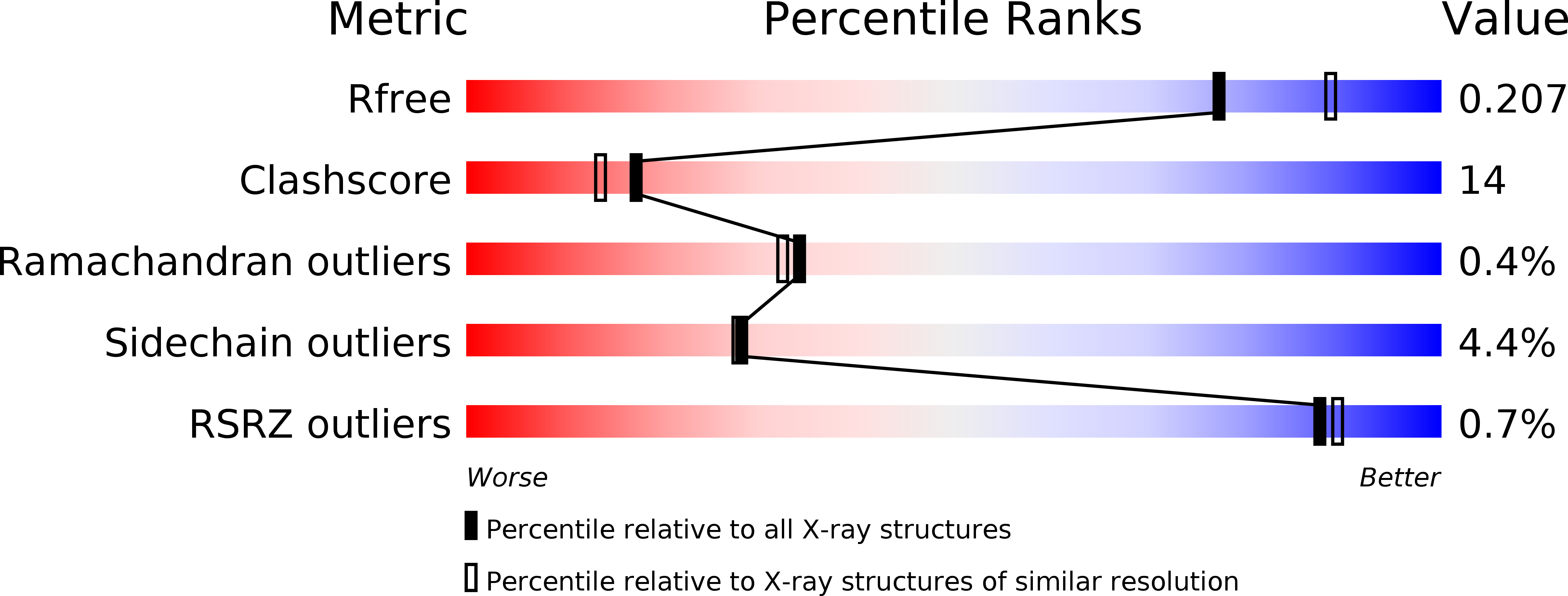
Structure of the PLP degradative enzyme 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase from Mesorhizobium loti MAFF303099 and its mechanistic implications.
McCulloch, K.M., Mukherjee, T., Begley, T.P., Ealick, S.E.(2009) Biochemistry 48: 4139-4149
- PubMed: 19317437
- DOI: https://doi.org/10.1021/bi900149f
- Primary Citation of Related Structures:
3GMB, 3GMC - PubMed Abstract:
A vitamin B(6) degradative pathway has recently been identified and characterized in Mesorhizobium loti MAFF303099. One of the enzymes on this pathway, 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase (MHPCO), is a flavin-dependent enzyme and catalyzes the oxidative ring-opening of 2-methyl-3-hydroxypyridine-5-carboxylic acid to form E-2-(acetamino-methylene)succinate. The gene for this enzyme has been cloned, and the corresponding protein has been overexpressed in Escherichia coli and purified. The crystal structure of MHPCO has been solved to 2.1 A using SAD phasing with and without the substrate MHPC bound. These crystal structures provide insight into the reaction mechanism and suggest roles for active site residues in the catalysis of a novel oxidative ring-opening reaction.
Organizational Affiliation:
Department of Chemistry and Chemical Biology, Cornell University, Ithaca, New York 14853, USA.