Rational development of high-affinity T-cell receptor-like antibodies
Stewart-Jones, G., Wadle, A., Hombach, A., Shenderov, E., Held, G., Fischer, E., Kleber, S., Stenner-Liewen, F., Bauer, S., McMichael, A., Knuth, A., Abken, H., Hombach, A.A., Cerundolo, V., Jones, E.Y., Renner, C.(2009) Proc Natl Acad Sci U S A 106: 5784-5788
- PubMed: 19307587
- DOI: https://doi.org/10.1073/pnas.0901425106
- Primary Citation of Related Structures:
3GJE, 3GJF, 3HAE - PubMed Abstract:
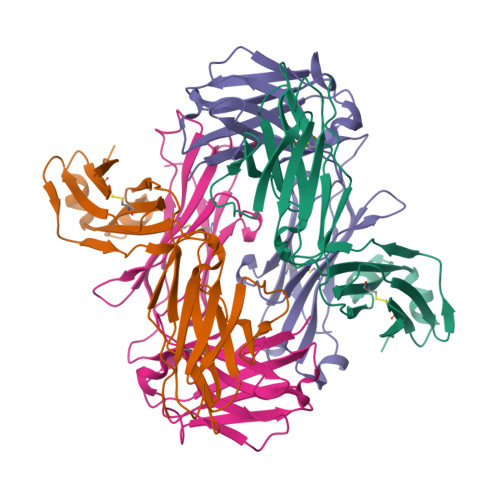
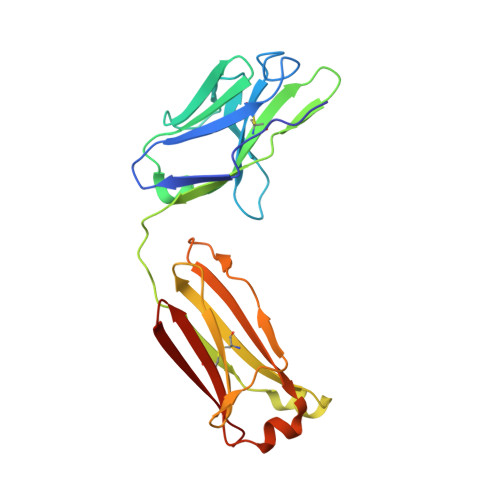
T-cell interaction with a target cell is a key event in the adaptive immune response and primarily driven by T-cell receptor (TCR) recognition of peptide-MHC (pMHC) complexes. TCR avidity for a given pMHC is determined by number of MHC molecules, availability of coreceptors, and TCR affinity for MHC or peptide, respectively, with peptide recognition being the most important factor to confer target specificity. Here we present high-resolution crystal structures of 2 Fab antibodies in complex with the immunodominant NY-ESO-1(157-165) peptide analogue (SLLMWITQV) presented by HLA-A*0201 and compare them with a TCR recognizing the same pMHC. Binding to the central methionine-tryptophan peptide motif and orientation of binding were almost identical for Fabs and TCR. As the MW "peg" dominates the contacts between Fab and peptide, we estimated the contributions of individual amino acids between the Fab and peptide to provide the rational basis for a peptide-focused second-generation, high-affinity antibody library. The final Fab candidate achieved better peptide binding by 2 light-chain mutations, giving a 20-fold affinity improvement to 2-4 nM, exceeding the affinity of the TCR by 1,000-fold. The high-affinity Fab when grafted as recombinant TCR on T cells conferred specific killing of HLA-A*0201/NY-ESO-1(157-165) target cells. In summary, we prove that affinity maturation of antibodies mimicking a TCR is possible and provide a strategy for engineering high-affinity antibodies that can be used in targeting specific pMHC complexes for diagnostic and therapeutic purposes.
Organizational Affiliation:
Division of Structural Biology, Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford OX3 7BN, United Kingdom.