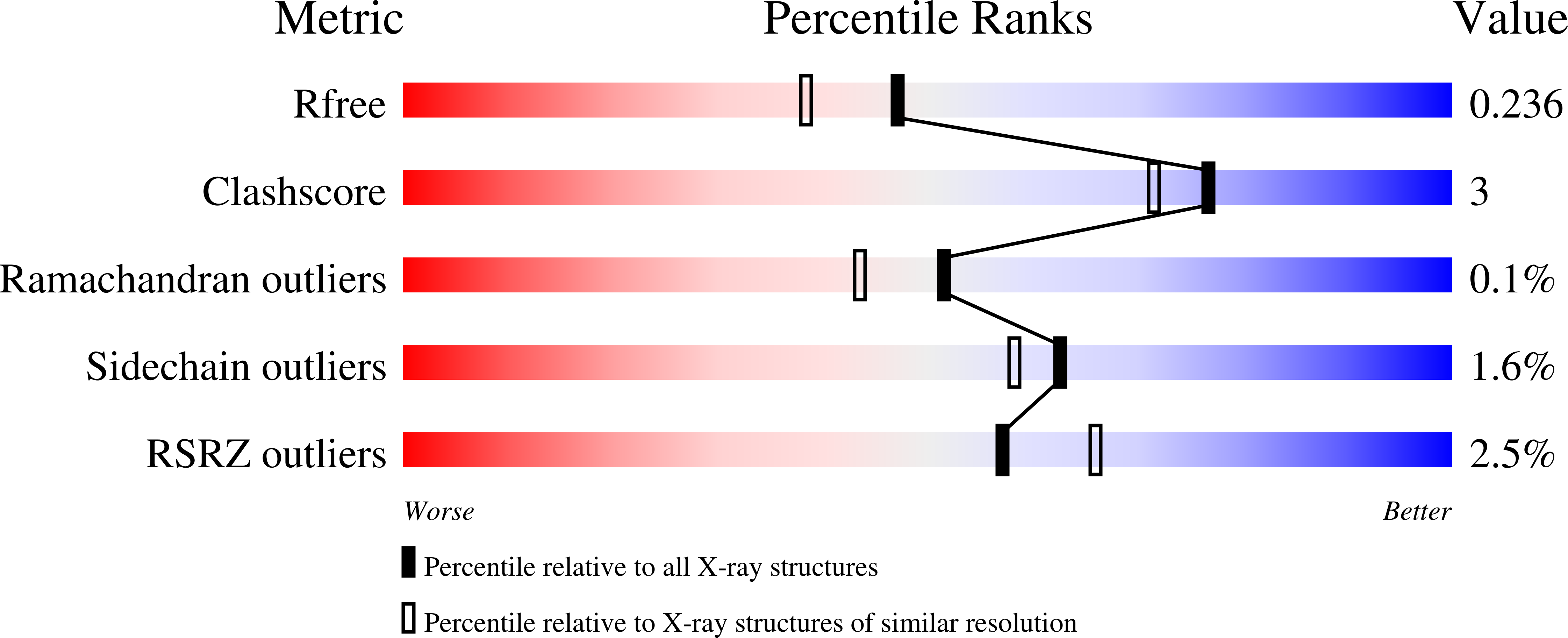
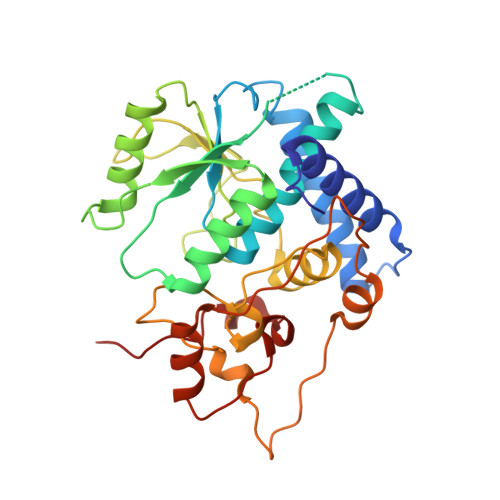
Structure and mechanism of a eukaryotic FMN adenylyltransferase.
Huerta, C., Borek, D., Machius, M., Grishin, N.V., Zhang, H.(2009) J Mol Biol 389: 388-400
- PubMed: 19375431
- DOI: https://doi.org/10.1016/j.jmb.2009.04.022
- Primary Citation of Related Structures:
3FWK, 3G59, 3G5A, 3G6K - PubMed Abstract:
Flavin mononucleotide adenylyltransferase (FMNAT) catalyzes the formation of the essential flavocoenzyme flavin adenine dinucleotide (FAD) and plays an important role in flavocoenzyme homeostasis regulation. By sequence comparison, bacterial and eukaryotic FMNAT enzymes belong to two different protein superfamilies and apparently utilize different sets of active-site residues to accomplish the same chemistry. Here we report the first structural characterization of a eukaryotic FMNAT from the pathogenic yeast Candida glabrata. Four crystal structures of C. glabrata FMNAT in different complexed forms were determined at 1.20-1.95 A resolutions, capturing the enzyme active-site states prior to and after catalysis. These structures reveal a novel flavin-binding mode and a unique enzyme-bound FAD conformation. Comparison of the bacterial and eukaryotic FMNATs provides a structural basis for understanding the convergent evolution of the same FMNAT activity from different protein ancestors. Structure-based investigation of the kinetic properties of FMNAT should offer insights into the regulatory mechanisms of FAD homeostasis by FMNAT in eukaryotic organisms.
Organizational Affiliation:
Department of Biochemistry, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA.