On the function and structure of synthetically modified porins.
Reitz, S., Cebi, M., Reiss, P., Studnik, G., Linne, U., Koert, U., Essen, L.O.(2009) Angew Chem Int Ed Engl 48: 4853-4857
- PubMed: 19322865
- DOI: https://doi.org/10.1002/anie.200900457
- Primary Citation of Related Structures:
3FYX - PubMed Abstract:
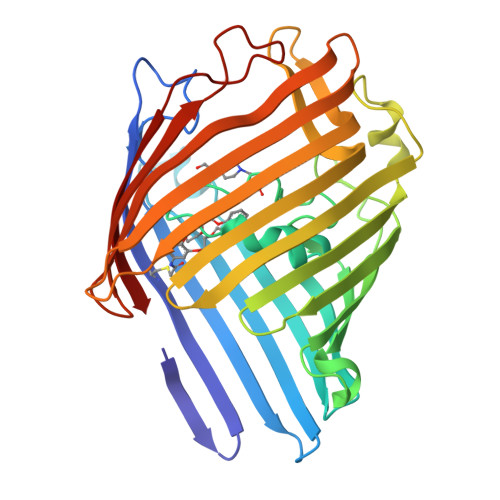
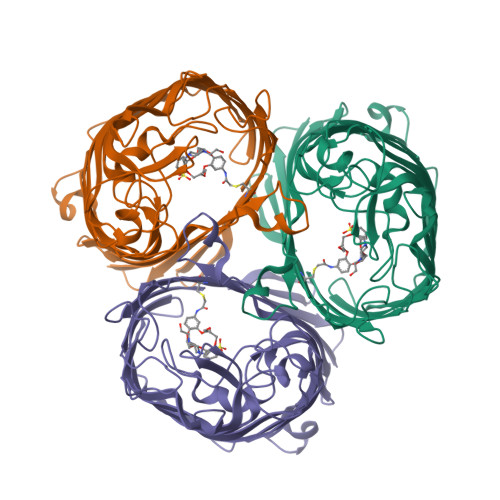
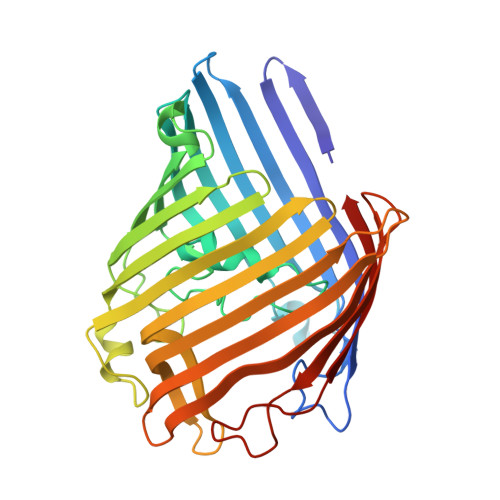
The attachment of modulators to a trimeric porin ion channel was investigated (see picture of the trimer with a crown ether modulator (orange)). The interplay of modulator and protein is essential for the conformational heterogeneity of the hybrid channel. Single-site attachment in large pores is not sufficient to change the electrophysiological characteristics of the pores-such change requires additional noncovalent interactions or second-site attachments.
Organizational Affiliation:
Fachbereich Chemie, Philipps-Universität Marburg, Hans-Meerwein-Strasse, 35032 Marburg, Germany.