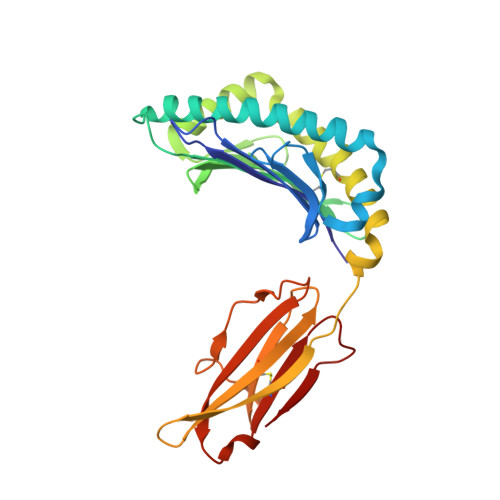
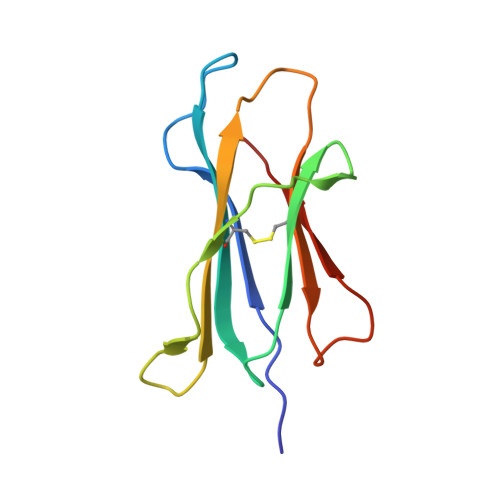
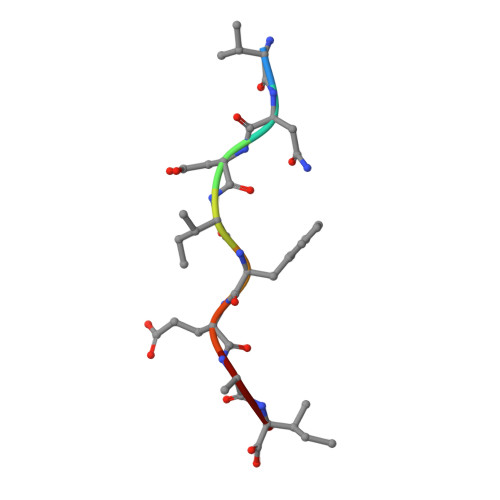
Predominant occupation of the class I MHC molecule H-2Kwm7 with a single self-peptide suggests a mechanism for its diabetes-protective effect.
Brims, D.R., Qian, J., Jarchum, I., Mikesh, L., Palmieri, E., Ramagopal, U.A., Malashkevich, V.N., Chaparro, R.J., Lund, T., Hattori, M., Shabanowitz, J., Hunt, D.F., Nathenson, S.G., Almo, S.C., Dilorenzo, T.P.(2010) Int Immunol 22: 191-203
- PubMed: 20093428
- DOI: https://doi.org/10.1093/intimm/dxp127
- Primary Citation of Related Structures:
3FOL, 3FOM, 3FON - PubMed Abstract:
Type 1 diabetes (T1D) is an autoimmune disease characterized by T cell-mediated destruction of insulin-producing pancreatic beta cells. In both humans and the non-obese diabetic (NOD) mouse model of T1D, class II MHC alleles are the primary determinant of disease susceptibility. However, class I MHC genes also influence risk. These findings are consistent with the requirement for both CD4(+) and CD8(+) T cells in the pathogenesis of T1D. Although a large body of work has permitted the identification of multiple mechanisms to explain the diabetes-protective effect of particular class II MHC alleles, studies examining the protective influence of class I alleles are lacking. Here, we explored this question by performing biochemical and structural analyses of the murine class I MHC molecule H-2K(wm7), which exerts a diabetes-protective effect in NOD mice. We have found that H-2K(wm7) molecules are predominantly occupied by the single self-peptide VNDIFERI, derived from the ubiquitous protein histone H2B. This unexpected finding suggests that the inability of H-2K(wm7) to support T1D development could be due, at least in part, to the failure of peptides from critical beta-cell antigens to adequately compete for binding and be presented to T cells. Predominant presentation of a single peptide would also be expected to influence T-cell selection, potentially leading to a reduced ability to select a diabetogenic CD8(+) T-cell repertoire. The report that one of the predominant peptides bound by T1D-protective HLA-A*31 is histone derived suggests the potential translation of our findings to human diabetes-protective class I MHC molecules.
Organizational Affiliation:
Department of Microbiology and Immunology, Albert Einstein College of Medicine, Bronx, NY 10461, USA.