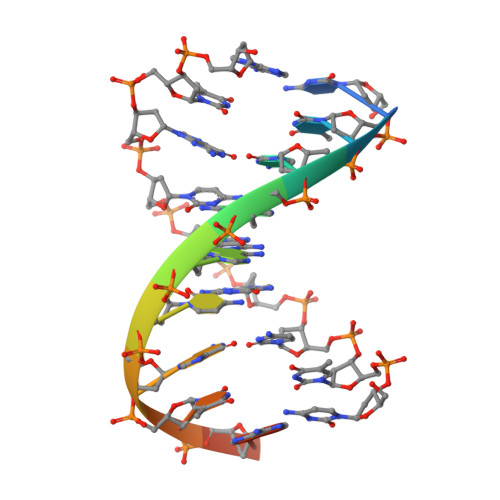
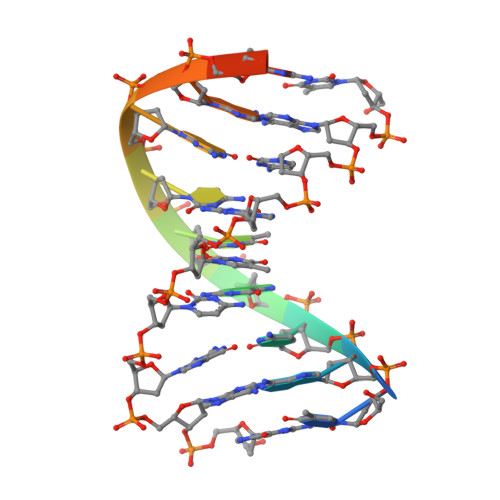
Crystal structure of the beta beta alpha-Me type II restriction endonuclease Hpy99I with target DNA.
Sokolowska, M., Czapinska, H., Bochtler, M.(2009) Nucleic Acids Res 37: 3799-3810
- PubMed: 19380375
- DOI: https://doi.org/10.1093/nar/gkp228
- Primary Citation of Related Structures:
3FC3, 3GOX - PubMed Abstract:
The beta beta alpha-Me restriction endonuclease (REase) Hpy99I recognizes the CGWCG target sequence and cleaves it with unusual stagger (five nucleotide 5'-recessed ends). Here we present the crystal structure of the specific complex of the dimeric enzyme with DNA. The Hpy99I protomer consists of an antiparallel beta-barrel and two beta 4 alpha 2 repeats. Each repeat coordinates a structural zinc ion with four cysteine thiolates in two CXXC motifs. The beta beta alpha-Me region of the second beta 4 alpha 2 repeat holds the catalytic metal ion (or its sodium surrogate) via Asp148 and Asn165 and activates a water molecule with the general base His149. In the specific complex, Hpy99I forms a ring-like structure around the DNA that contacts DNA bases on the major and minor groove sides via the first and second beta 4 alpha 2 repeats, respectively. Hpy99I interacts with the central base pair of the recognition sequence only on the minor groove side, where A:T resembles T:A and G:C is similar to C:G. The Hpy99I-DNA co-crystal structure provides the first detailed illustration of the beta beta alpha-Me site in REases and complements structural information on the use of this active site motif in other groups of endonucleases such as homing endonucleases (e.g. I-PpoI) and Holliday junction resolvases (e.g. T4 endonuclease VII).
Organizational Affiliation:
International Institute of Molecular and Cell Biology, Trojdena 4, 02-109 Warsaw, Poland.