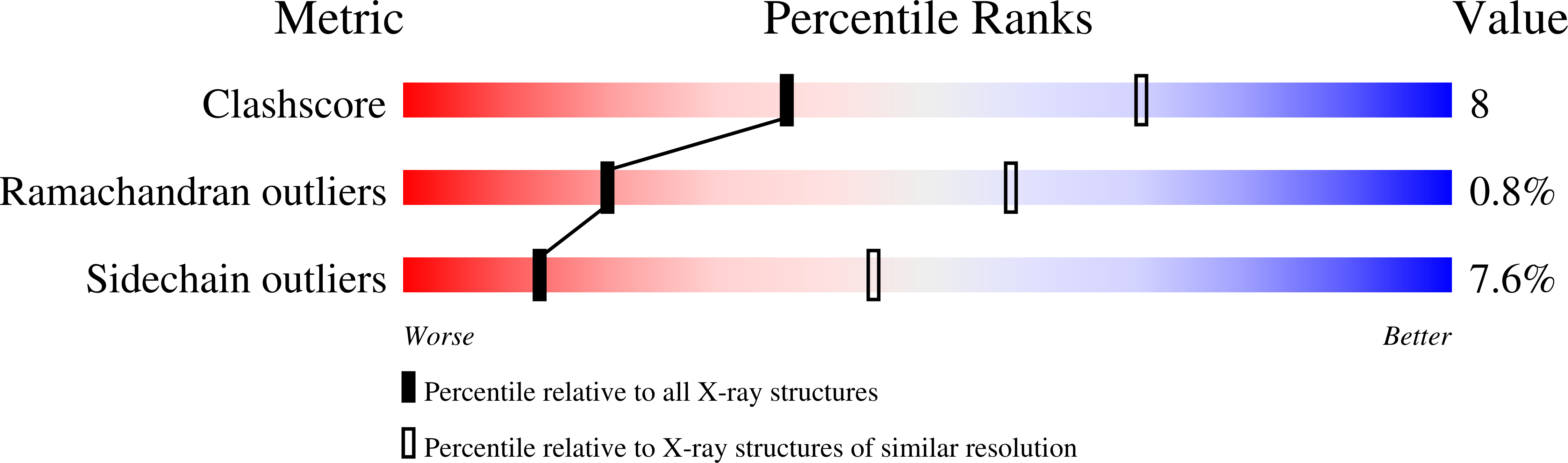Structures of the multicomponent Rieske non-heme iron toluene 2,3-dioxygenase enzyme system
Friemann, R., Lee, K., Brown, E.N., Gibson, D.T., Eklund, H., Ramaswamy, S.(2009) Acta Crystallogr D Biol Crystallogr 65: 24-33
- PubMed: 19153463
- DOI: https://doi.org/10.1107/S0907444908036524
- Primary Citation of Related Structures:
3DQY, 3EF6, 3EN1, 3EQQ - PubMed Abstract:
Bacterial Rieske non-heme iron oxygenases catalyze the initial hydroxylation of aromatic hydrocarbon substrates. The structures of all three components of one such system, the toluene 2,3-dioxygenase system, have now been determined. This system consists of a reductase, a ferredoxin and a terminal dioxygenase. The dioxygenase, which was cocrystallized with toluene, is a heterohexamer containing a catalytic and a structural subunit. The catalytic subunit contains a Rieske [2Fe-2S] cluster and mononuclear iron at the active site. This iron is not strongly bound and is easily removed during enzyme purification. The structures of the enzyme with and without mononuclear iron demonstrate that part of the structure is flexible in the absence of iron. The orientation of the toluene substrate in the active site is consistent with the regiospecificity of oxygen incorporation seen in the product formed. The ferredoxin is Rieske type and contains a [2Fe-2S] cluster close to the protein surface. The reductase belongs to the glutathione reductase family of flavoenzymes and consists of three domains: an FAD-binding domain, an NADH-binding domain and a C-terminal domain. A model for electron transfer from NADH via FAD in the reductase and the ferredoxin to the terminal active-site mononuclear iron of the dioxygenase is proposed.
Organizational Affiliation:
Department of Molecular Biology, Swedish University of Agricultural Sciences, Box 590, 75124 Uppsala, Sweden.