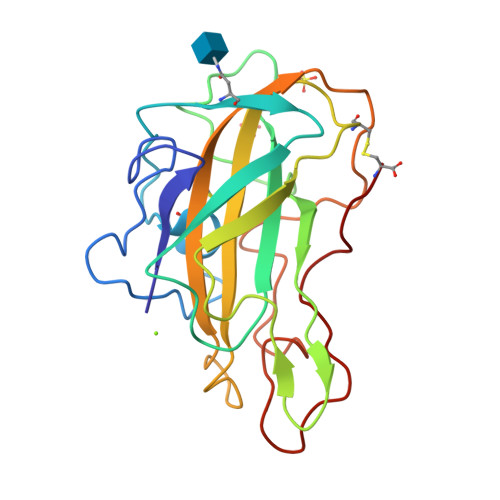
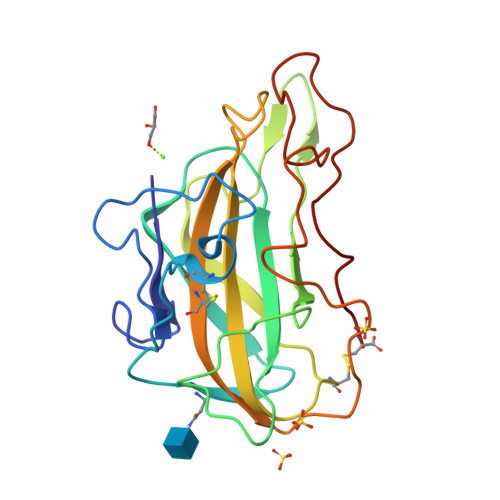
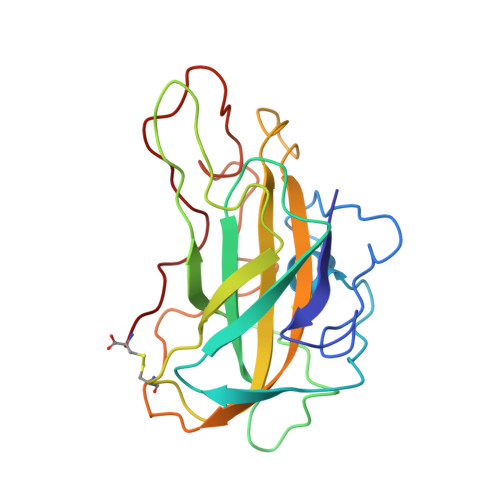
Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family.
Harris, P.V., Welner, D., McFarland, K.C., Re, E., Navarro Poulsen, J.C., Brown, K., Salbo, R., Ding, H., Vlasenko, E., Merino, S., Xu, F., Cherry, J., Larsen, S., Lo Leggio, L.(2010) Biochemistry 49: 3305-3316
- PubMed: 20230050
- DOI: https://doi.org/10.1021/bi100009p
- Primary Citation of Related Structures:
3EII, 3EJA - PubMed Abstract:
Currently, the relatively high cost of enzymes such as glycoside hydrolases that catalyze cellulose hydrolysis represents a barrier to commercialization of a biorefinery capable of producing renewable transportable fuels such as ethanol from abundant lignocellulosic biomass. Among the many families of glycoside hydrolases that catalyze cellulose and hemicellulose hydrolysis, few are more enigmatic than family 61 (GH61), originally classified based on measurement of very weak endo-1,4-beta-d-glucanase activity in one family member. Here we show that certain GH61 proteins lack measurable hydrolytic activity by themselves but in the presence of various divalent metal ions can significantly reduce the total protein loading required to hydrolyze lignocellulosic biomass. We also solved the structure of one highly active GH61 protein and find that it is devoid of conserved, closely juxtaposed acidic side chains that could serve as general proton donor and nucleophile/base in a canonical hydrolytic reaction, and we conclude that the GH61 proteins are unlikely to be glycoside hydrolases. Structure-based mutagenesis shows the importance of several conserved residues for GH61 function. By incorporating the gene for one GH61 protein into a commercial Trichoderma reesei strain producing high levels of cellulolytic enzymes, we are able to reduce by 2-fold the total protein loading (and hence the cost) required to hydrolyze lignocellulosic biomass.
Organizational Affiliation:
Novozymes Inc., 1445 Drew Avenue, Davis, California 95618, USA. paha@novozymes.com