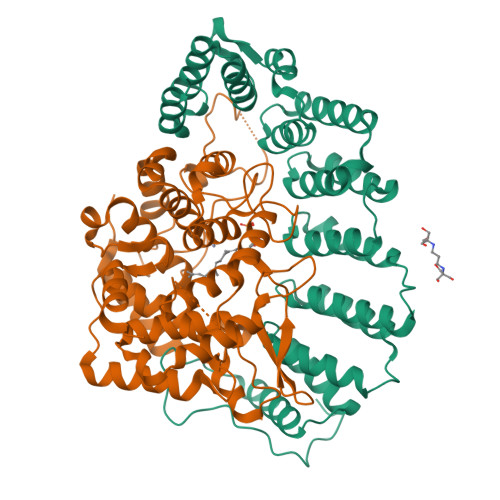
Structure of protein geranylgeranyltransferase-I from the human pathogen Candida albicans complexed with a lipid substrate.
Hast, M.A., Beese, L.S.(2008) J Biological Chem 283: 31933-31940
- PubMed: 18713740
- DOI: https://doi.org/10.1074/jbc.M805330200
- Primary Citation of Related Structures:
3DRA - PubMed Abstract:
Protein geranylgeranyltransferase-I (GGTase-I) catalyzes the transfer of a 20-carbon isoprenoid lipid to the sulfur of a cysteine residue located near the C terminus of numerous cellular proteins, including members of the Rho superfamily of small GTPases and other essential signal transduction proteins. In humans, GGTase-I and the homologous protein farnesyltransferase (FTase) are targets of anticancer therapeutics because of the role small GTPases play in oncogenesis. Protein prenyltransferases are also essential for many fungal and protozoan pathogens that infect humans, and have therefore become important targets for treating infectious diseases. Candida albicans, a causative agent of systemic fungal infections in immunocompromised individuals, is one pathogen for which protein prenylation is essential for survival. Here we present the crystal structure of GGTase-I from C. albicans (CaGGTase-I) in complex with its cognate lipid substrate, geranylgeranylpyrophosphate. This structure provides a high-resolution picture of a non-mammalian protein prenyltransferase. There are significant variations between species in critical areas of the active site, including the isoprenoid-binding pocket, as well as the putative product exit groove. These differences indicate the regions where specific protein prenyltransferase inhibitors with antifungal activity can be designed.
Organizational Affiliation:
Department of Biochemistry, Duke University Medical Center, Durham, North Carolina 27710, USA.