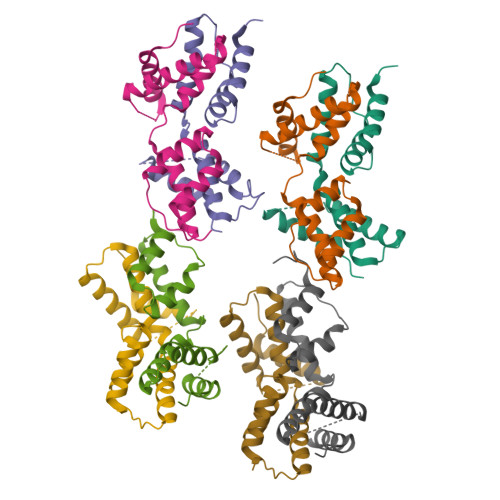
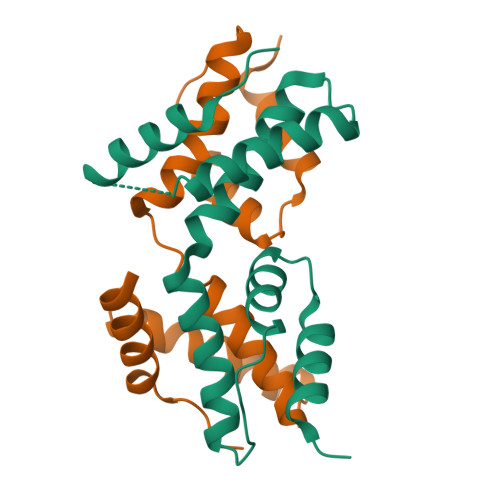
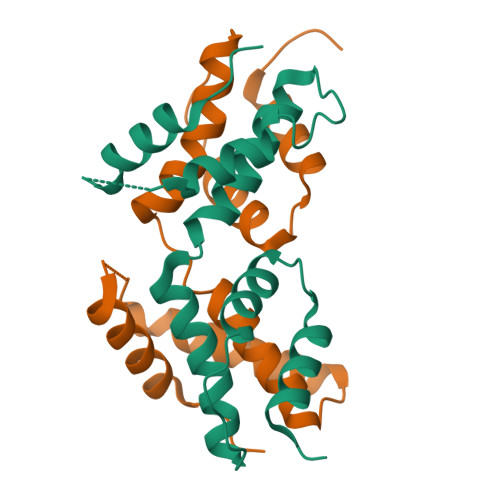
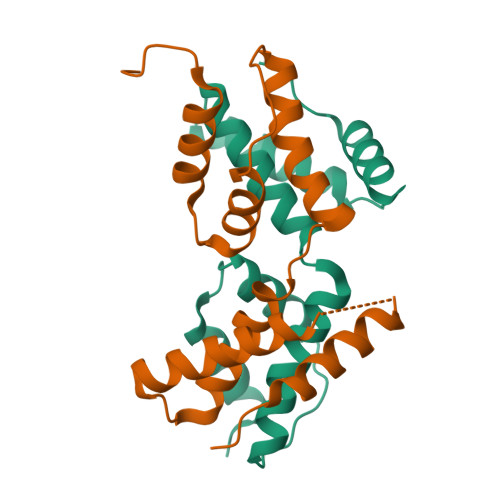
The intrinsically disordered domain of the antitoxin Phd chaperones the toxin Doc against irreversible inactivation and misfolding
De Gieter, S., Konijnenberg, A., Talavera, A., Butterer, A., Haesaerts, S., De Greve, H., Sobott, F., Loris, R., Garcia-Pino, A.(2014) J Biological Chem 289: 34013-34023
- PubMed: 25326388
- DOI: https://doi.org/10.1074/jbc.M114.572396
- Primary Citation of Related Structures:
3DD9 - PubMed Abstract:
The toxin Doc from the phd/doc toxin-antitoxin module targets the cellular translation machinery and is inhibited by its antitoxin partner Phd. Here we show that Phd also functions as a chaperone, keeping Doc in an active, correctly folded conformation. In the absence of Phd, Doc exists in a relatively expanded state that is prone to dimerization through domain swapping with its active site loop acting as hinge region. The domain-swapped dimer is not capable of arresting protein synthesis in vitro, whereas the Doc monomer is. Upon binding to Phd, Doc becomes more compact and is secured in its monomeric state with a neutralized active site.
Organizational Affiliation:
From Structural Biology Brussels, Department of Biotechnology (DBIT), Vrije Universiteit Brussel, Pleinlaan 2, B-1050 Brussels, Belgium, Molecular Recognition Unit (MoRe).