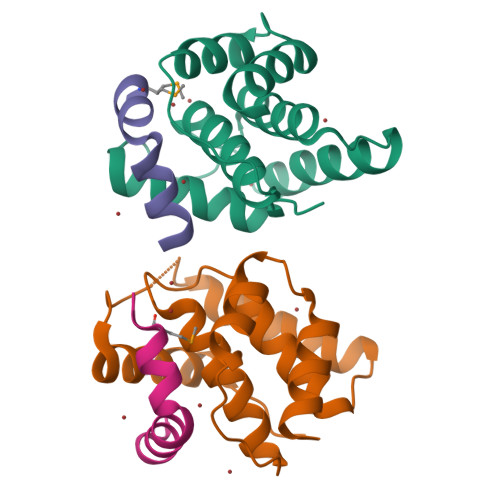
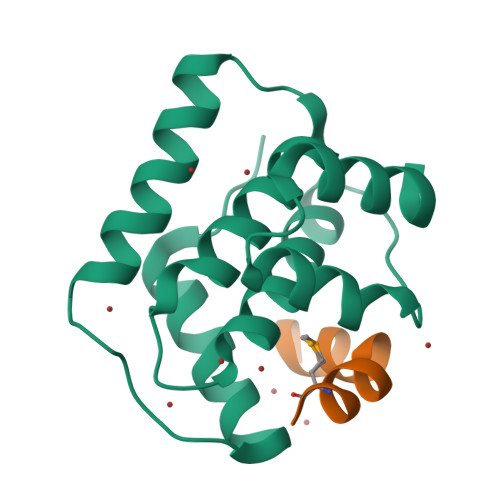
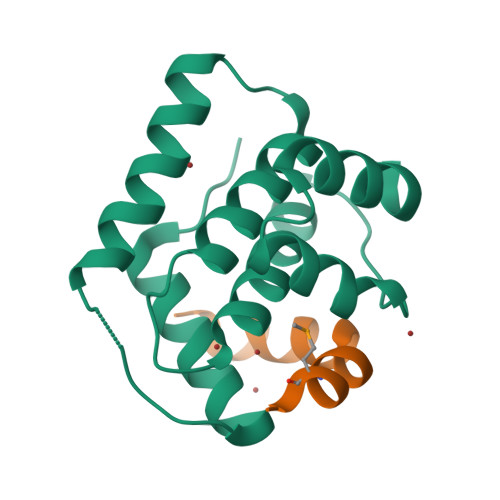
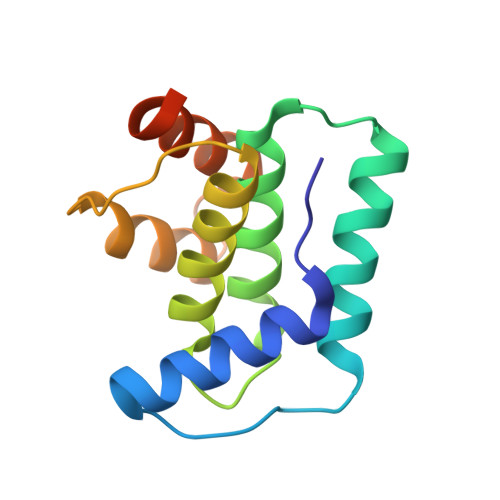
Doc of Prophage P1 Is Inhibited by Its Antitoxin Partner Phd through Fold Complementation
Garcia-Pino, A., Christensen-Dalsgaard, M., Wyns, L., Yarmolinsky, M., Magnuson, R.D., Gerdes, K., Loris, R.(2008) J Biological Chem 283: 30821-30827
- PubMed: 18757857
- DOI: https://doi.org/10.1074/jbc.M805654200
- Primary Citation of Related Structures:
3DD7 - PubMed Abstract:
Prokaryotic toxin-antitoxin modules are involved in major physiological events set in motion under stress conditions. The toxin Doc (death on curing) from the phd/doc module on phage P1 hosts the C-terminal domain of its antitoxin partner Phd (prevents host death) through fold complementation. This Phd domain is intrinsically disordered in solution and folds into an alpha-helix upon binding to Doc. The details of the interactions reveal the molecular basis for the inhibitory action of the antitoxin. The complex resembles the Fic (filamentation induced by cAMP) proteins and suggests a possible evolutionary origin for the phd/doc operon. Doc induces growth arrest of Escherichia coli cells in a reversible manner, by targeting the protein synthesis machinery. Moreover, Doc activates the endogenous E. coli RelE mRNA interferase but does not require this or any other known chromosomal toxin-antitoxin locus for its action in vivo.
Organizational Affiliation:
Laboratorium voor Ultrastructuur, Vrije Universiteit Brussel, Pleinlaan 2, B-1050 Brussel, Belgium.