Crystal structures of TAFI elucidate the inactivation mechanism of activated TAFI: a novel mechanism for enzyme autoregulation
Marx, P.F., Brondijk, T.H., Plug, T., Romijn, R.A., Hemrika, W., Meijers, J.C.M., Huizinga, E.G.(2008) Blood 112: 2803-2809
- PubMed: 18559974
- DOI: https://doi.org/10.1182/blood-2008-03-146001
- Primary Citation of Related Structures:
3D66, 3D67, 3D68 - PubMed Abstract:
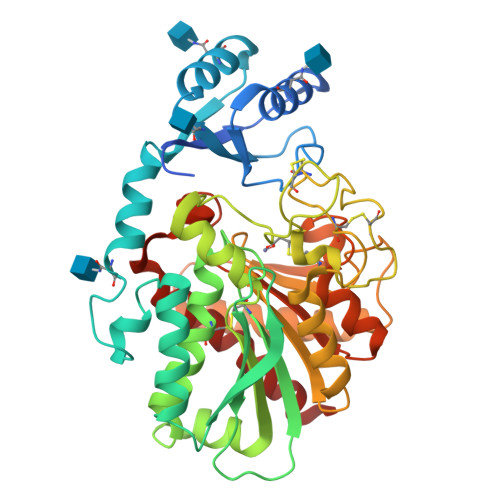
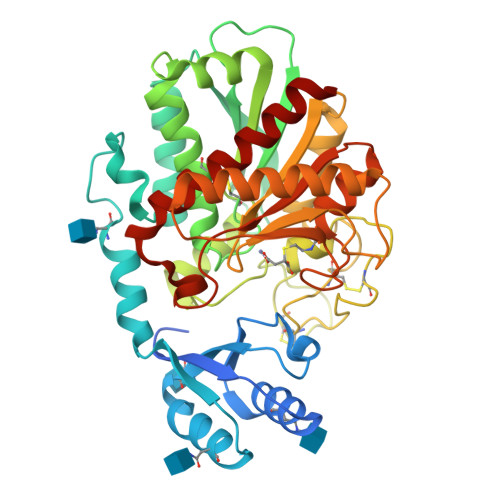
Thrombin-activatable fibrinolysis inhibitor (TAFI) is a pro-metallocarboxypeptidase that can be proteolytically activated (TAFIa). TAFIa is unique among carboxypeptidases in that it spontaneously inactivates with a short half-life, a property that is crucial for its role in controlling blood clot lysis. We studied the intrinsic instability of TAFIa by solving crystal structures of TAFI, a TAFI inhibitor (GEMSA) complex and a quadruple TAFI mutant (70-fold more stable active enzyme). The crystal structures show that TAFIa stability is directly related to the dynamics of a 55-residue segment (residues 296-350) that includes residues of the active site wall. Dynamics of this flap are markedly reduced by the inhibitor GEMSA, a known stabilizer of TAFIa, and stabilizing mutations. Our data provide the structural basis for a model of TAFI auto-regulation: in zymogen TAFI the dynamic flap is stabilized by interactions with the activation peptide. Release of the activation peptide increases dynamic flap mobility and in time this leads to conformational changes that disrupt the catalytic site and expose a cryptic thrombin-cleavage site present at Arg302. This represents a novel mechanism of enzyme control that enables TAFI to regulate its activity in plasma in the absence of specific inhibitors.
Organizational Affiliation:
Department of Experimental Vascular Medicine, Academic Medical Center, University of Amsterdam, Amsterdam, The Netherlands.