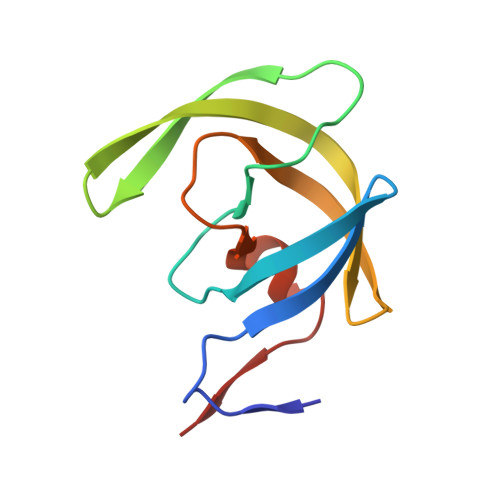
Structural analysis of human immunodeficiency virus type 1 CRF01_AE protease in complex with the substrate p1-p6.
Bandaranayake, R.M., Prabu-Jeyabalan, M., Kakizawa, J., Sugiura, W., Schiffer, C.A.(2008) J Virol 82: 6762-6766
- PubMed: 18434392
- DOI: https://doi.org/10.1128/JVI.00018-08
- Primary Citation of Related Structures:
3D3T - PubMed Abstract:
The effect of amino acid variability between human immunodeficiency virus type 1 (HIV-1) clades on structure and the emergence of resistance mutations in HIV-1 protease has become an area of significant interest in recent years. We determined the first crystal structure of the HIV-1 CRF01_AE protease in complex with the p1-p6 substrate to a resolution of 2.8 A. Hydrogen bonding between the flap hinge and the protease core regions shows significant structural rearrangements in CRF01_AE protease compared to the clade B protease structure.
Organizational Affiliation:
Department of Biochemistry and Molecular Pharmacology, University of Massachusetts Medical School, 364 Plantation Street, Worcester, MA 01605, USA.