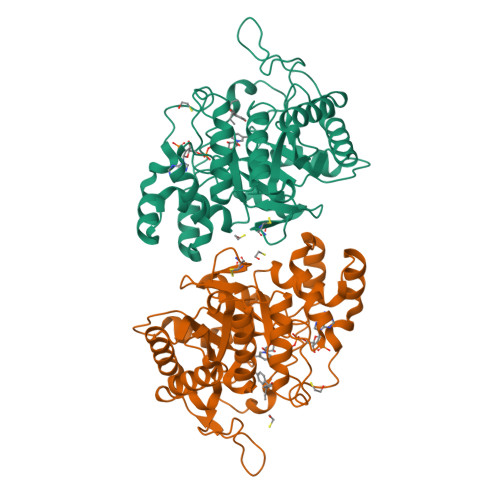
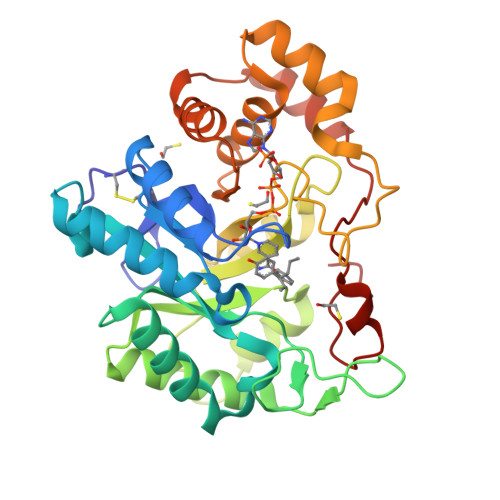
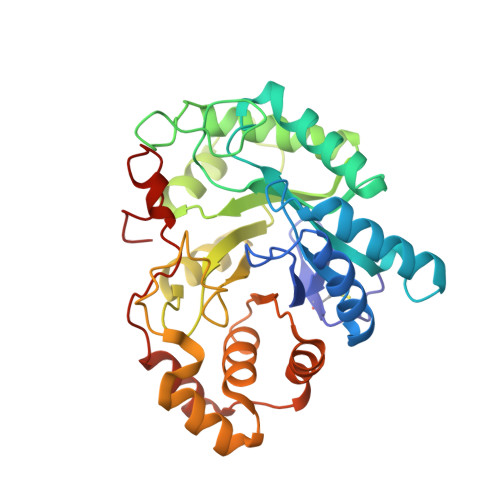
Structure of the G225P/G226P mutant of mouse 3(17)alpha-hydroxysteroid dehydrogenase (AKR1C21) ternary complex: implications for the binding of inhibitor and substrate.
Dhagat, U., Endo, S., Mamiya, H., Hara, A., El-Kabbani, O.(2009) Acta Crystallogr D Biol Crystallogr 65: 257-265
- PubMed: 19237748
- DOI: https://doi.org/10.1107/S0907444908044028
- Primary Citation of Related Structures:
3CV6 - PubMed Abstract:
3(17)alpha-Hydroxysteroid dehydrogenase (AKR1C21) is a unique member of the aldo-keto reductase (AKR) superfamily owing to its ability to reduce 17-ketosteroids to 17alpha-hydroxysteroids, as opposed to other members of the AKR family, which can only produce 17beta-hydroxysteroids. In this paper, the crystal structure of a double mutant (G225P/G226P) of AKR1C21 in complex with the coenzyme NADP(+) and the inhibitor hexoestrol refined at 2.1 A resolution is presented. Kinetic analysis and molecular-modelling studies of 17alpha- and 17beta-hydroxysteroid substrates in the active site of AKR1C21 suggested that Gly225 and Gly226 play an important role in determining the substrate stereospecificity of the enzyme. Additionally, the G225P/G226P mutation of the enzyme reduced the affinity (K(m)) for both 3alpha- and 17alpha-hydroxysteroid substrates by up to 160-fold, indicating that these residues are critical for the binding of substrates.
Organizational Affiliation:
Medicinal Chemistry and Drug Action, Monash Institute of Pharmaceutical Sciences, Monash University, Parkville, Victoria 3052, Australia.