The FACT Spt16 "peptidase" domain is a histone H3-H4 binding module
Stuwe, T., Hothorn, M., Lejeune, E., Rybin, V., Bortfeld, M., Scheffzek, K., Ladurner, A.G.(2008) Proc Natl Acad Sci U S A 105: 8884-8889
- PubMed: 18579787
- DOI: https://doi.org/10.1073/pnas.0712293105
- Primary Citation of Related Structures:
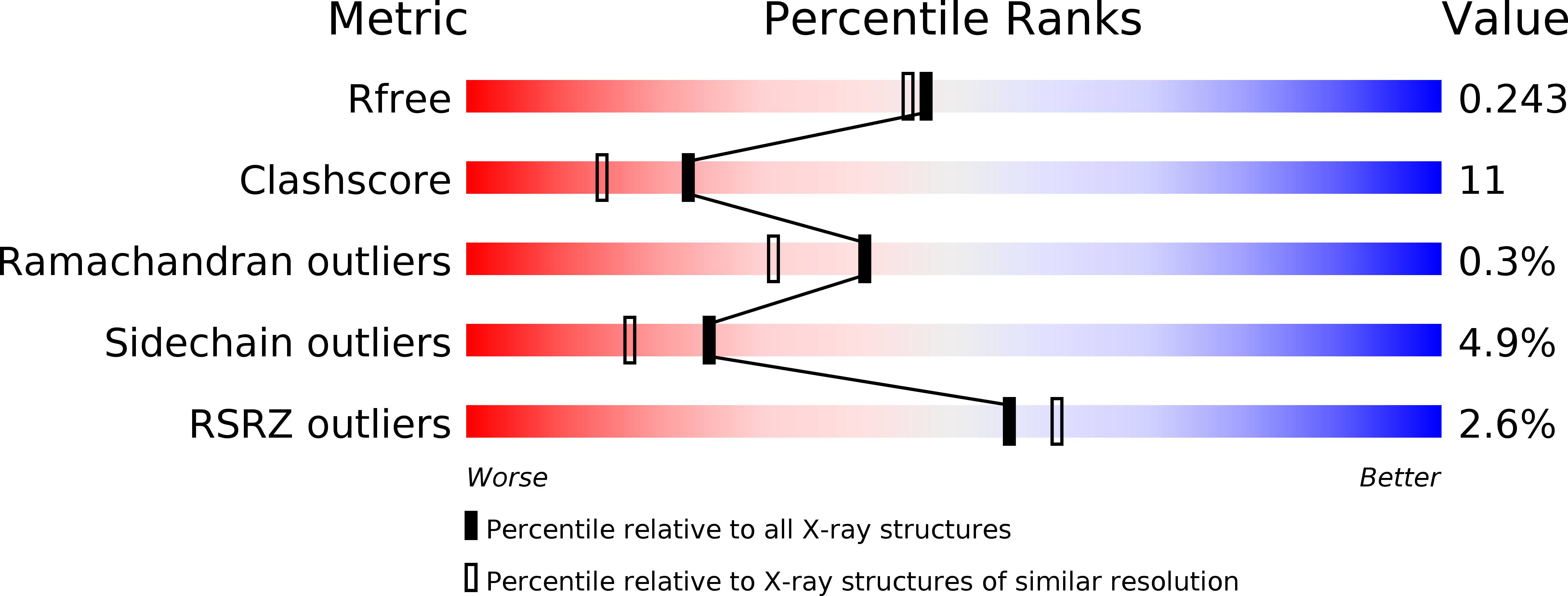
3CB5, 3CB6 - PubMed Abstract:
The FACT complex is a conserved cofactor for RNA polymerase II elongation through nucleosomes. FACT bears histone chaperone activity and contributes to chromatin integrity. However, the molecular mechanisms behind FACT function remain elusive. Here we report biochemical, structural, and mutational analyses that identify the peptidase homology domain of the Schizosaccharomyces pombe FACT large subunit Spt16 (Spt16-N) as a binding module for histones H3 and H4. The 2.1-A crystal structure of Spt16-N reveals an aminopeptidase P fold whose enzymatic activity has been lost. Instead, the highly conserved fold directly binds histones H3-H4 through a tight interaction with their globular core domains, as well as with their N-terminal tails. Mutations within a conserved surface pocket in Spt16-N or posttranslational modification of the histone H4 tail reduce interaction in vitro, whereas the globular domains of H3-H4 and the H3 tail bind distinct Spt16-N surfaces. Our analysis suggests that the N-terminal domain of Spt16 may add to the known H2A-H2B chaperone activity of FACT by including a H3-H4 tail and H3-H4 core binding function mediated by the N terminus of Spt16. We suggest that these interactions may aid FACT-mediated nucleosome reorganization events.
Organizational Affiliation:
European Molecular Biology Laboratory, Meyerhofstrasse 1, 69117 Heidelberg, Germany.