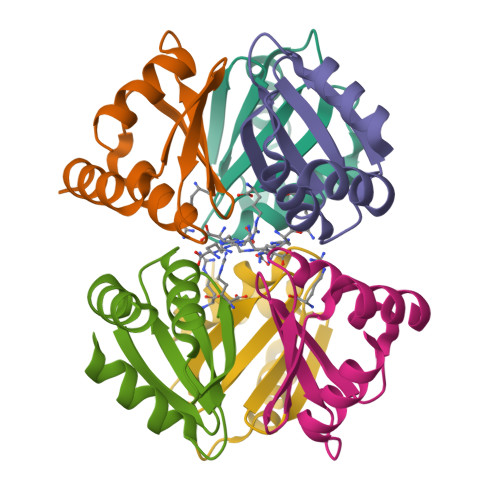
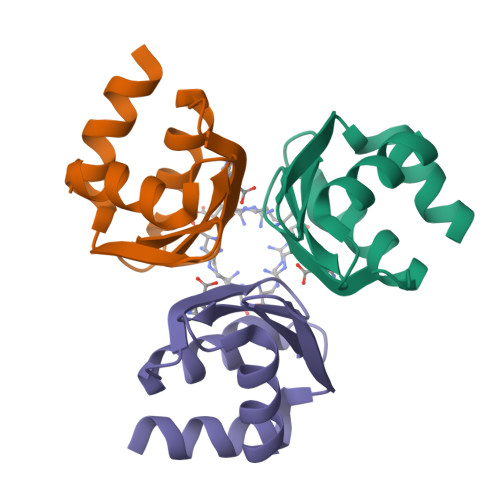
Structure of the C-terminal domain of the arginine repressor protein from Mycobacterium tuberculosis.
Cherney, L.T., Cherney, M.M., Garen, C.R., Lu, G.J., James, M.N.(2008) Acta Crystallogr D Biol Crystallogr 64: 950-956
- PubMed: 18703843
- DOI: https://doi.org/10.1107/S0907444908021513
- Primary Citation of Related Structures:
2ZFZ, 3BUE, 3CAG - PubMed Abstract:
The Mycobacterium tuberculosis (Mtb) gene product encoded by open reading frame Rv1657 is an arginine repressor (ArgR). All genes involved in the L-arginine (hereafter arginine) biosynthetic pathway are essential for optimal growth of the Mtb pathogen, thus making MtbArgR a potential target for drug design. The C-terminal domains of arginine repressors (CArgR) participate in oligomerization and arginine binding. Several crystal forms of CArgR from Mtb (MtbCArgR) have been obtained. The X-ray crystal structures of MtbCArgR were determined at 1.85 A resolution with bound arginine and at 2.15 A resolution in the unliganded form. These structures show that six molecules of MtbCArgR are arranged into a hexamer having approximate 32 point symmetry that is formed from two trimers. The trimers rotate relative to each other by about 11 degrees upon binding arginine. All residues in MtbCArgR deemed to be important for hexamer formation and for arginine binding have been identified from the experimentally determined structures presented. The hexamer contains six regular sites in which the arginine molecules have one common binding mode and three sites in which the arginine molecules have two overlapping binding modes. The latter sites only bind the ligand at high (200 mM) arginine concentrations.
Organizational Affiliation:
Group in Protein Structure and Function, Department of Biochemistry, University of Alberta, Edmonton, Alberta T6G 2H7, Canada.