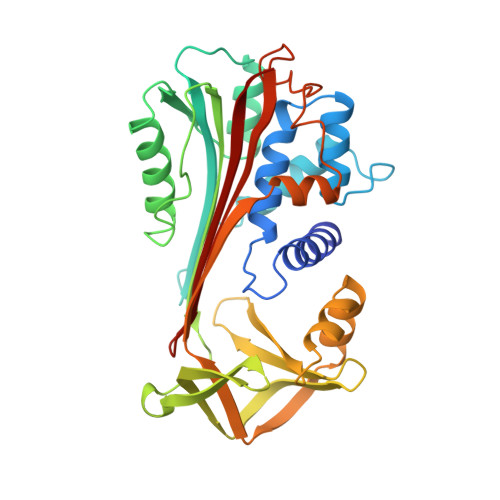
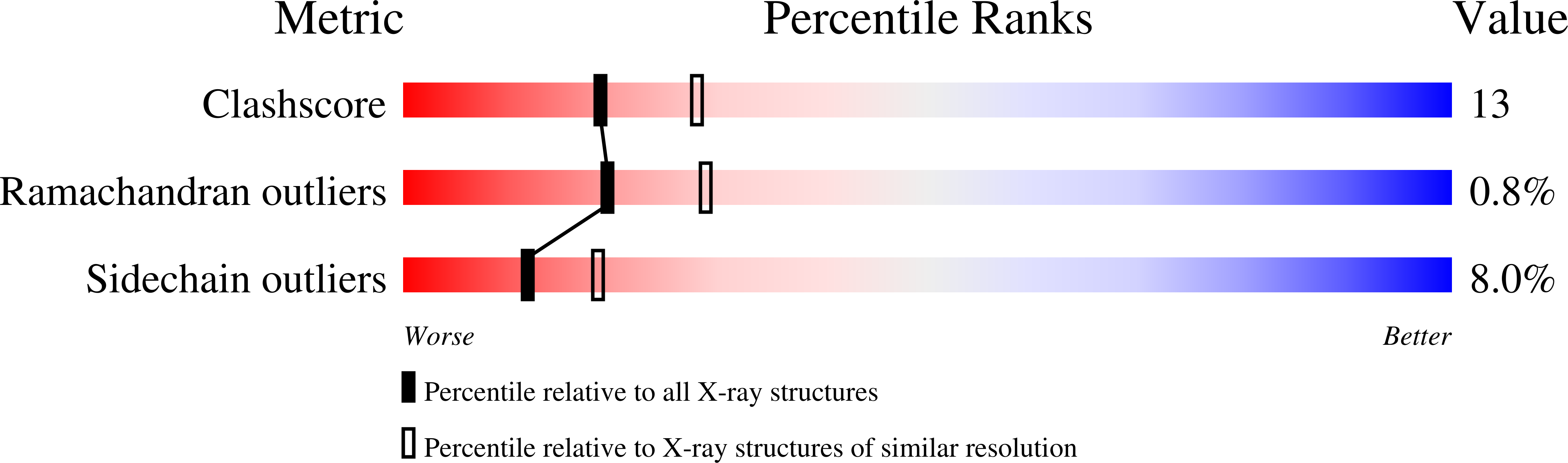Engineering an anion-binding cavity in antichymotrypsin modulates the "spring-loaded" serpin-protease interaction.
Lukacs, C.M., Rubin, H., Christianson, D.W.(1998) Biochemistry 37: 3297-3304
- PubMed: 9521649
- DOI: https://doi.org/10.1021/bi972359e
- Primary Citation of Related Structures:
1AS4, 3CAA, 4CAA - PubMed Abstract:
Expressed in a kinetically trapped folding state, a serpin couples the thermodynamic driving force of a massive beta-sheet rearrangement to the inhibition of a target protease. Hence, the serpin-protease interaction is the premier example of a "spring-loaded" protein-protein interaction. Amino acid substitutions in the hinge region of a serpin reactive loop can weaken the molecular spring, which converts the serpin from an inhibitor into a substrate. To probe the molecular basis of this conversion, we report the crystal structure of A349R antichymotrypsin in the reactive loop cleaved state at 2.1 A resolution. This amino acid substitution does not block the beta-sheet rearrangement despite the burial of R349 in the hydrophobic core of the cleaved serpin along with a salt-linked acetate ion. The inhibitory activity of this serpin variant is not obliterated; remarkably, its inhibitory properties are anion-dependent due to the creation of an anion-binding cavity in the cleaved serpin.
Organizational Affiliation:
Department of Chemistry, University of Pennsylvania, Philadelphia 19104, USA.